- Title
-
Macrophage Reprogramming with Anti-miR223-Loaded Artificial Protocells Enhances In Vivo Cancer Therapeutic Potential
- Authors
- López-Cuevas, P., Xu, C., Severn, C.E., Oates, T.C.L., Cross, S.J., Toye, A.M., Mann, S., Martin, P.
- Source
- Full text @ Adv Sci (Weinh)
|
miR223 inhibition reduces cancer progression in zebrafish. A) Pigmentation of the tailfin quantified by threshold analysis of tail region (red dotted outline) in 1‐month‐old cancerous Ras;WT versus Ras;miR223KO juvenile fish. B) Dot plot showing percentage of pigmentation quantified from the regions imaged in (A) and (1). C) Ras‐GFP expression in the tail area (red dotted outline) of 8‐month‐old cancerous Ras;WT versus equivalent Ras;miR223KO adult fish. D) Dot plot showing levels of Ras‐GFP expression as quantified by fluorescent pixel count (FPC) from the regions imaged in (C) and (2). E) 1‐year‐old cancerous Ras;WT versus Ras;miR223KO adult fish bearing (or not) a tumor mass (red outline) on their tail. F) Bar chart showing percentage of cancerous fish with or without a tail tumor quantified from the regions imaged in (E) and (3). G) Dot plot showing tumor area quantified from the regions imaged in (E) and (3). H) Bar chart showing percentage of cancerous Ras;WT versus Ras;miR223KO adult fish bearing tailfin tumors, and any additional tumor (or not), quantified from the regions imaged in (4)–(6). See also Figure |
|
Systemically injected free‐circulating protocells are taken up by leukocytes. A) Schematic representation of dextran‐containing FITC‐labeled proteinosome‐based protocells. B) Single‐channel confocal image of FITC‐protocells. C) Multi‐channel confocal image of the flank of a 2 dpf casper larva after systemic injection of FITC‐protocells at 0.5 hpi; white arrows indicate the direction of the blood flow. D) High magnification view of (C) showing FITC‐protocells moving through the caudal vein at 24 hpi; red blood cells are also imaged (asterisks). See also Figure |
|
Locally injected protocells are taken up by macrophages. A) Multi‐channel confocal images of the flank of Tg( |
|
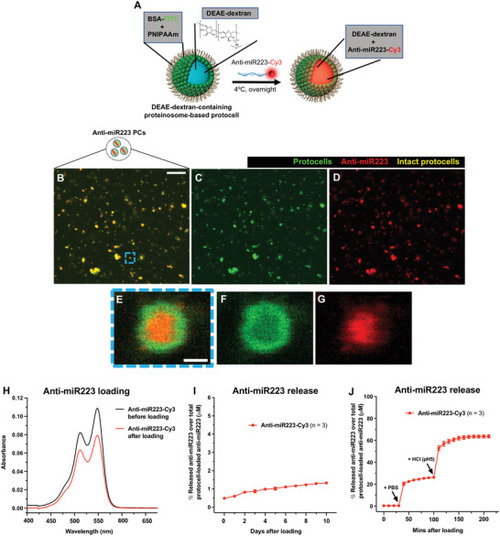
Loading strategy of anti‐miR223 into protocells. A) Schematic of the experimental design for loading DEAE‐dextran‐containing FITC‐protocells with anti‐miR223‐Cy3. B–G) Multi‐channel (B,E) or single‐channel (C,D,F,G) confocal images of FITC‐protocells after loading with anti‐miR223‐Cy3. E–G) High magnification views of (B)–(D) showing a single anti‐miR223‐Cy3 FITC‐protocell. H) Spectra showing absorbance quantification of the anti‐miR223‐Cy3 supernatant before and after protocell loading. I,J) Graphs showing percentage of anti‐miR223‐Cy3 released from loaded protocells into the supernatant over total anti‐miR223‐Cy3 concentration initially loaded into protocells, under normal conditions in H2O (I) or after exposure to PBS and HCl (J). Data are representative (H) or pooled (I,J) from three independent experiments. Graphs (I,J) show mean ± SEM, and each dot represents the mean of all experiments analyzed. |
|
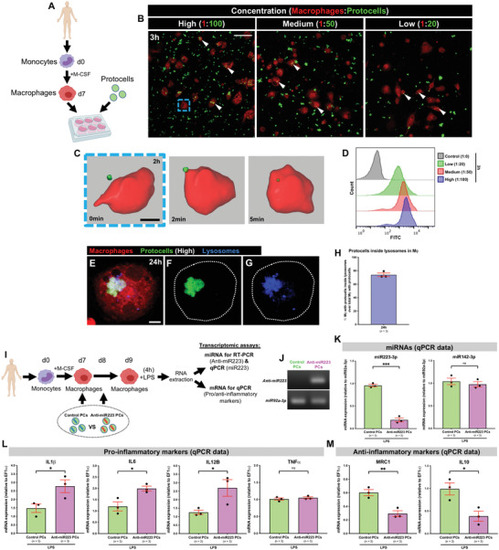
Uptake of anti‐miR223 protocells enhances il1 |
|
In vitro uptake of anti‐miR223 protocells by human macrophages enhances expression of pro‐inflammatory markers. A) Schematic of the experimental design; monocytes isolated from human donors undergo in vitro differentiation toward macrophages (via M‐CSF) prior to FITC‐protocell addition (day 7). B) Multi‐channel confocal images of human macrophages after incubation with different FITC‐protocell concentrations for 3 h; white arrowheads indicate protocells within macrophages. C) Imaris 3D reconstruction from confocal movie frames showing FITC‐protocell uptake by a human macrophage 2 h after protocell addition (blue dashed box in [B]). See also Movie |
|
Uptake of anti‐miR223 protocells induces a leukocyte pro‐inflammatory state in adult zebrafish. A) Multi‐channel image of a 1‐year‐old adult tail tumor (red outline) locally injected with FITC‐protocells and imaged at 0.5 hpi. B) High magnification view of (A) showing a single‐channel image of FITC‐protocells (white arrowheads) at the injection site. C) Multi‐channel confocal image of an immunostained cryosection from a 1‐year‐old adult tail tumor harvested 6 h after local FITC‐protocell injection; nuclei are stained with DAPI (blue), and leukocytes are revealed by anti‐L‐plastin immunostaining (red). D) High magnification view of (C) showing single‐channel confocal image of Ras‐GFP region. E–I) High magnification views of (C) showing multi‐channel (E–H) or single‐channel (I) confocal images of FITC‐protocells within L‐plastin‐positive cells (white dotted outline in [I]). See also Figure |
|
Anti‐miR223 protocell treatment reduces cancer progression in adult zebrafish. A–H) Single‐channel confocal images of immunostained cryosections from 1‐year‐old adult tail tumors at 3 dpt after a single local injection of unlabeled control protocells (A,B,E,F) or unlabeled anti‐miR223 protocells (C,D,G,H); white lines indicate tumor margins; proliferating cells are revealed by anti‐pH3 immunostaining (magenta) in (A)–(D) and apoptotic cells revealed by TUNEL staining (yellow) in (E)–(H). B,D,F,H) High magnification views of (A), (C), (E), and (G). I,J) Graphs showing percentage of pH3‐positive (I) or TUNEL‐positive (J) area over total tumor area from sections after each injection regime quantified from the regions imaged in (A), (C), (E), and (G). K–R) Multi‐channel (K,M,O,Q) or single‐channel (L,N,P,R) images of 1‐year‐old adult tail tumors (red outlines) at 0 and 30 dpt after multiple local injections of unlabeled control protocells (K–N) or unlabeled anti‐miR223 protocells (O–R). S) Graph showing adult tail tumor growth curves after each protocell treatment quantified from the regions imaged in (K)‐(R) and (1). See also Figure |