- Title
-
Zebrafish as a Model to Unveil the Pro-Osteogenic Effects of Boron-Vitamin D3 Synergism
- Authors
- Sojan, J.M., Gundappa, M.K., Carletti, A., Gaspar, V., Gavaia, P., Maradonna, F., Carnevali, O.
- Source
- Full text @ Front Nutr
|
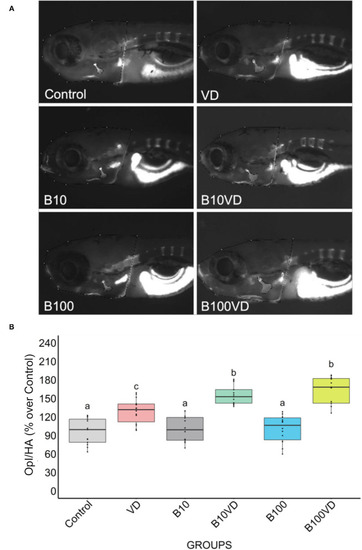
Fluorescence microphotograph of AR-S-stained larvae and ImageJ quantification of the opercular bone mineralization. |
|
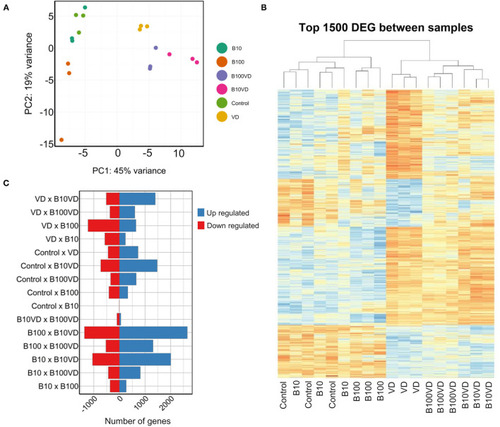
Sample clustering and DEGs across different combinations. |
|
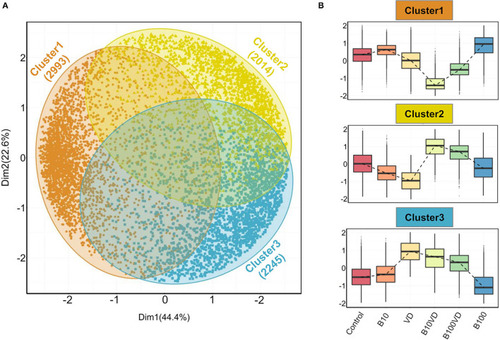
Outputs from PAM clustering of all the DEGs. |
|
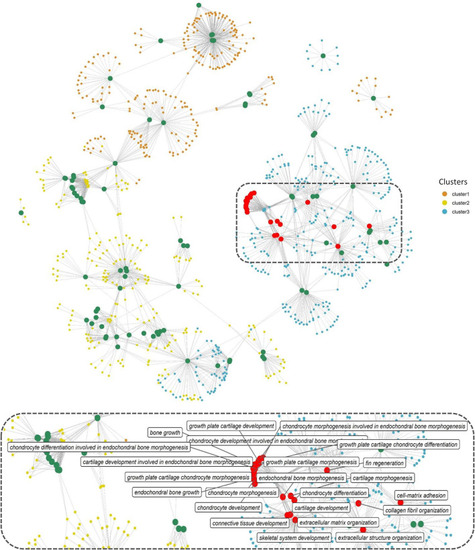
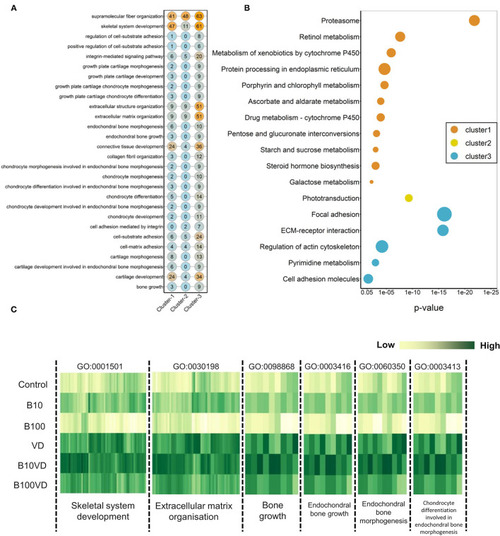
Network representation of enriched GO terms (BP) performed against the DEGs across three different clusters ( |
|
|
|
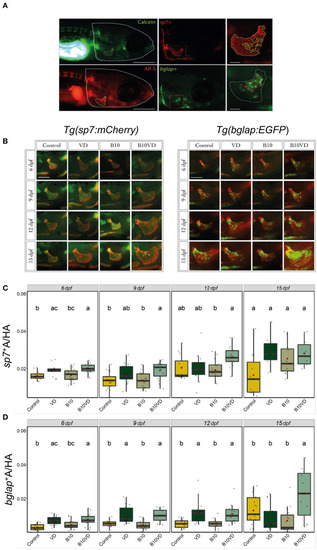
Early and mature osteoblasts respectively tracked by the fluorescence expression of |