- Title
-
Generation and Characterization of a Zebrafish IL-2Rγc SCID Model
- Authors
- Sertori, R., Jones, R., Basheer, F., Rivera, L., Dawson, S., Loke, S., Heidary, S., Dhillon, A., Liongue, C., Ward, A.C.
- Source
- Full text @ Int. J. Mol. Sci.
|
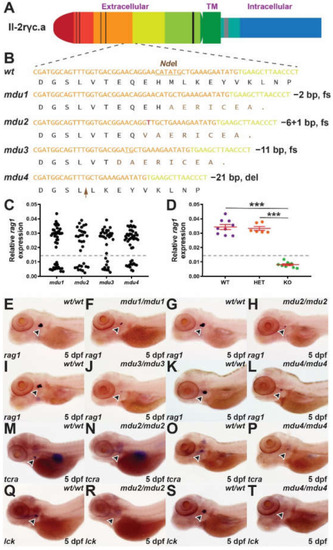
Identification of zebrafish il-2rγc.a mutant alleles and their impact on embryonic lymphopoiesis. (A) Schematic of zebrafish Il-2rγc.a with the extracellular domain, containing cysteine bonds (thin black lines) and a WSXWS motif (thick black line), shown to the left, followed by the transmembrane (TM) domain and the intracellular domain containing Box 1 (thick grey line), with alternate coloring depicting derivation from different exons. (B) Mutant il2rga alleles. Sequence of wild-type (wt) and indicated mutant (mdu) alleles, il2rgamdu1, il2rgamdu2, il2rgamdu3, and il2rgamdu4, along with their impacts at the nucleotide and protein level (fs, frameshift; del, deletion). Nucleotide sequences are colored according to their derivation from exon 3 (orange), exon 4 (yellow), or de novo (red), with the NdeI cut site used for genotyping underlined. The corresponding protein translations are shown below, with wild-type sequence in black and that derived from an alternative reading frame in brown, with the position of the seven residue deletion in mdu4 indicated with the brown arrow. (C–T) WISH analysis. Embryos obtained from in-crossing of heterozygotes carrying the indicated il2rga alleles were fixed at 5 dpf and subjected to WISH with the indicated markers. Expression of rag1 in individual embryos was quantified relative to eye size for each allele (C) (n = 31–46), and a selection of embryos genotyped by NdeI digestion to allow determination of their genotype (WT, wild-type; HET, heterozygote; KO homozygote mutant) (D), showing results for individual embryos, with mean and SEM in red and statistical significance indicated (***: p < 0.001, n = 6–9). Expression of rag1 (indicated with arrowheads) in representative wild-type and homozygous mutant embryos derived from il2rgamdu1 (mdu1) (E,F), il2rgamdu2 (mdu2) (G,H), il2rgamdu3 (mdu3) (I,J) and il2rgamdu4 (mdu4) (K,L), tcra from il2rgamdu2 (M,N) and il2rgamdu4 (O,P) and lck from il2rgamdu2 (Q,R) and il2rgamdu4 (S,T) are shown. |
|
Effect of Il-2rγc.a ablation on later hematopoiesis. (A,B) Gene expression analysis. Larvae (28 dpf) obtained from in-crossing il2rgawt/mdu2 heterozygotes were harvested for gDNA and total RNA. The gDNA was used for the identification of wild-type (WT), heterozygote (HET), and homozygote knock-out (KO) siblings, and the RNA was subjected to RT-PCR with primers specific for T cell receptor β chain V(D)J-Cβ rearrangements (tcrb: vb1.5, vb12, vb14.5), B cell immunoglobulin M heavy chain gene rearrangements (ighm: vh1, vh4), and β-actin (actb) as a control (A) RT-negative controls yielded no products (data not shown) (n = 2). (B) Adult kidney derived total RNA was subjected to qRT2-PCR with gene markers of T cells (cd4, tcra), B cells (ighm, cd79a), NK cells (nklb), neutrophils (mpo), and monocyte/macrophages (mpeg1.1) (B) Data was normalized relative to actb and represented as relative fold change compared to wild-type fish, with mean and SD shown in red and statistical significance indicated (**: p < 0.01, ***: p < 0.001, n = 6). Histological analysis (C–H) Representative images of Giemsa-stained kidney (C,D) and blood (E,F) cells were obtained from WT (C,E) and KO (D,F) fish. Abbreviations: e, erythrocyte; l, lymphocyte; n, neutrophil; p, precursor. Differential counts of kidney (G) and blood (H) cells, showing results for individual embryos, with mean and SD in red and statistical significance indicated (*: p < 0.05, **: p < 0.01, ***: p < 0.001, kidney: n = 11–12, blood: n = 6). EXPRESSION / LABELING:
PHENOTYPE:
|
|
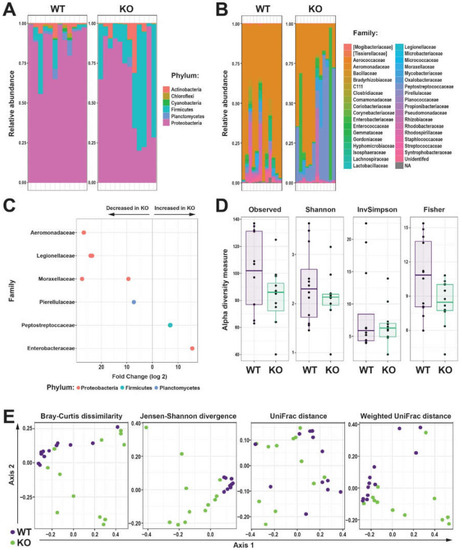
Effect of Il-2rγc.a ablation on intestinal microbiota. (A,B) Relative abundance of bacterial Amplicon sequence variants (ASVs) identified to the taxonomic level of phylum (A) and family (B) in the intestinal microbiome of individual WT (purple) and KO (green) zebrafish. (C) Differential abundances of bacterial family statistically different (p < 0.05) between WT and KO zebrafish, with the direction of change indicated. (D) Alpha diversity of WT and KO zebrafish intestinal microbiome displaying Observed, Shannon, InvSimpson, and Fisher’s indices as box and whisker plots. (E) Beta diversity of WT (purple) and KO (green) zebrafish intestinal microbiome displaying Bray-Curtis dissimilarity, Jensen-Shannon divergence, Unifrac, and Weighted Unifrac of log-transformed relative abundances ordination plots, with each dot representing an individual fish (n = 12 each genotype). |
|
Effect of Il-2rγc.a ablation on tumor immunity. (A–H) Representative casper fish harboring il2rgawt/wt (casper wild-type) (A,C,E,G) or il2rgamdu2/mdu2 (casper scid) (B,D,F,H) injected with fluorescently-labeled human HCT116 colorectal cancer cells imaged at the indicated days post injection (dpi). (I–I’. Histology of a representative tumor from a casper scid fish at 18 dpi at low magnification (5×, I) and of the boxed area at high magnification (40×, J). (K) Quantitation of tumor area showing results for individual embryos, with mean and SEM in red. (L) Kaplan–Meier survival curve of transplanted fish. For panels (K–L), statistical significance is indicated (**: p < 0.01, ***: p < 0.001, n = 11–24). |