- Title
-
Co-occurrence of BAP1 and SF3B1 mutations in uveal melanoma induces cellular senescence
- Authors
- Yu, L., Zhou, D., Zhang, G., Ren, Z., Luo, X., Liu, P., Plouffe, S.W., Meng, Z., Moroishi, T., Li, Y., Zhang, Y., Brown, J.H., Liu, S., Guan, K.L.
- Source
- Full text @ Mol. Oncol.
|
BRCA1‐associated protein 1 (BAP1) deficiency in UM cells with mutant splicing factor 3B subunit 1 ( |
|
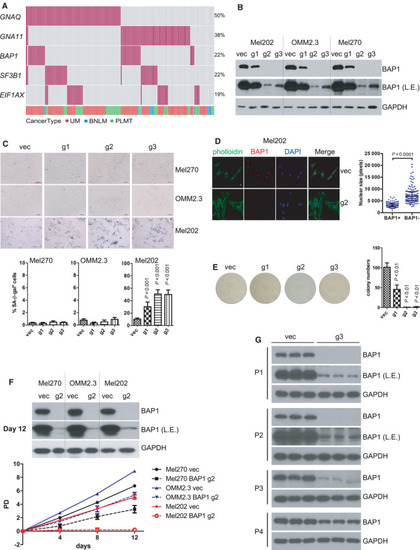
BAP1 deficiency combined with SF3B1 hotspot mutation induces senescence. (A) Growth curve of Mel202 cells with different |
|
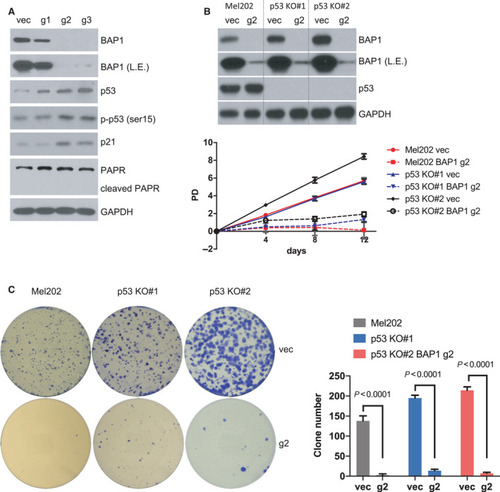
p53 is not required for the senescence phenotype. (A) Immunoblots show the expression of indicated proteins of Mel202 cells infected by vector control (vec) or |
|
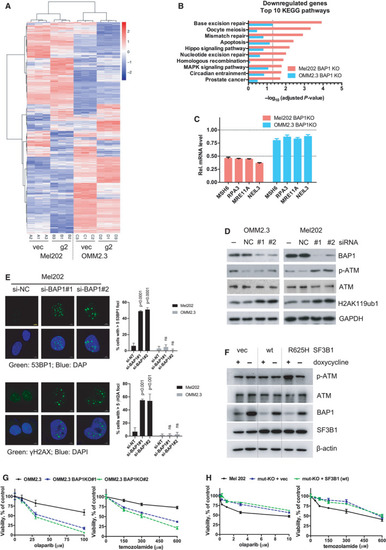
BAP1 deficiency combined with SF3B1 hotspot mutation induces DNA damage response. (A) RNA‐Seq data of differentially expressed genes (DEGs) in Mel202 and OMM2.3 cells with BAP1 KO (g2) compared with vector control (vec). Data were analyzed with DESeq2 (cutoff: adjusted |
|
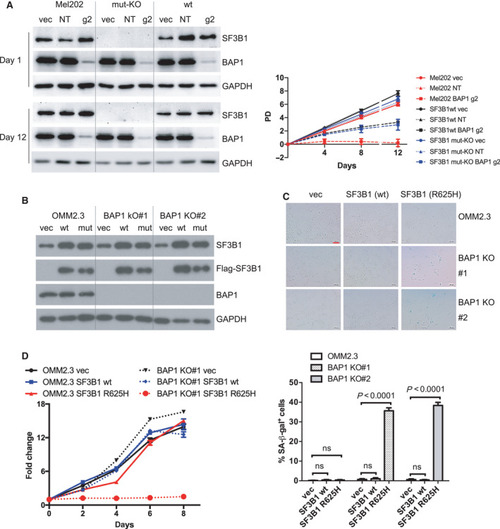
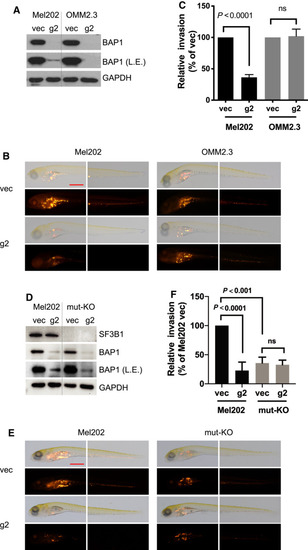
BAP1 deletion suppresses |
|
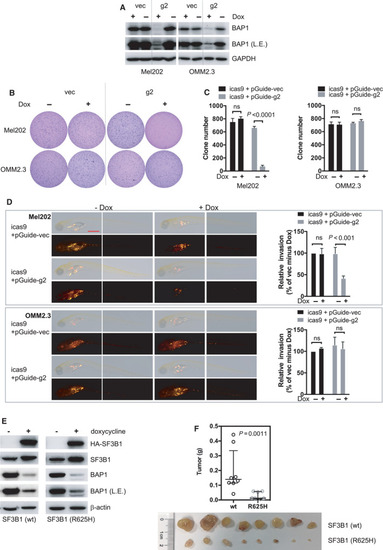
BAP1 deficiency combined with SF3B1 mutation suppresses in vivo invasion and growth of UM cells. (A) Immunoblots show BAP1 KO efficacy by inducible lentivirus CRIPSR‐Cas9. Stable Mel202 and OMM2.3 cell clones with Cas9 expression under doxycycline‐inducible promoter were selected. These Cas9‐expressing clones were then infected with lentivirus encoding for control vector (vec) or BAP1‐targeting sgRNA (g2). Cells were treated with doxycycline (200 ng·mL−1) for 6 days. The blots shown are representative of three independent experiments. (B) Soft‐agar colony‐formation assay of Mel202 and OMM2.3 cells upon inducible BAP1 deletion. Colonies were stained with crystal violet. The images shown are representative of three independent experiments. (C) Quantification of colonies. The data are presented as the mean ± SD (n = 3) from one representative experiment out of three. Student’s t test was used for statistical analysis. (D) Representative images at 6 dpi of parental and inducible CRISPR‐mediated BAP1 knockout Mel202 and OMM2.3 heterogeneous cell pool engrafted into the yolk sac of zebrafish embryos. The scale bar represents 500 μm. After injection, embryos were soaked in egg water containing doxycycline (10 μg·mL−1). Graphs show quantification of tumor cell invasion. All experiments consist of three independent repeats. For each independent experiment, at least 10 embryos were used in each group. Values of three independent experiments were given as mean ± SD. Student’s t test was used for statistical analysis. (E) Immunoblots show inducible BAP1 knockout and inducible expression of wild‐type or mutant SF3B1 (R625H) in OCM1 cells. Cells were treated with doxycycline (200 ng·mL−1) for 3 days, and BAP1 deletion and wild‐type/mutant SF3B1 were assessed by immunoblotting. The blots shown are representative of three independent experiments. (F) BAP1 knockout combined with mutant SF3B1 expression inhibits OCM1 tumor xenograft growth in vivo. The same cell lines used in (E) were grafted into nude mice subcutaneously, and tumor weight was quantified. Data are presented as median with interquartile range. n = 8 mice per group. The Mann–Whitney U‐test was used for statistical analysis. |
|
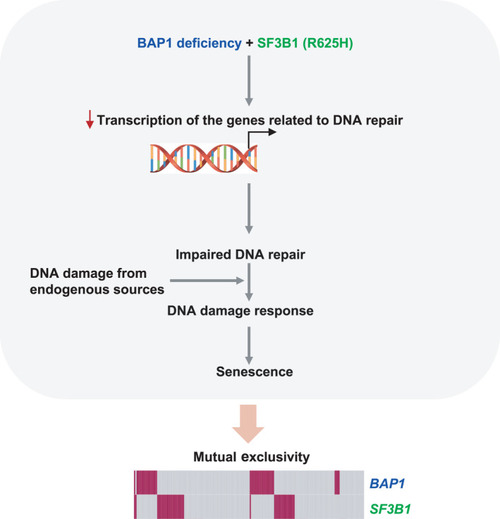
A proposed model accounting for the observed mutually exclusive pattern of |