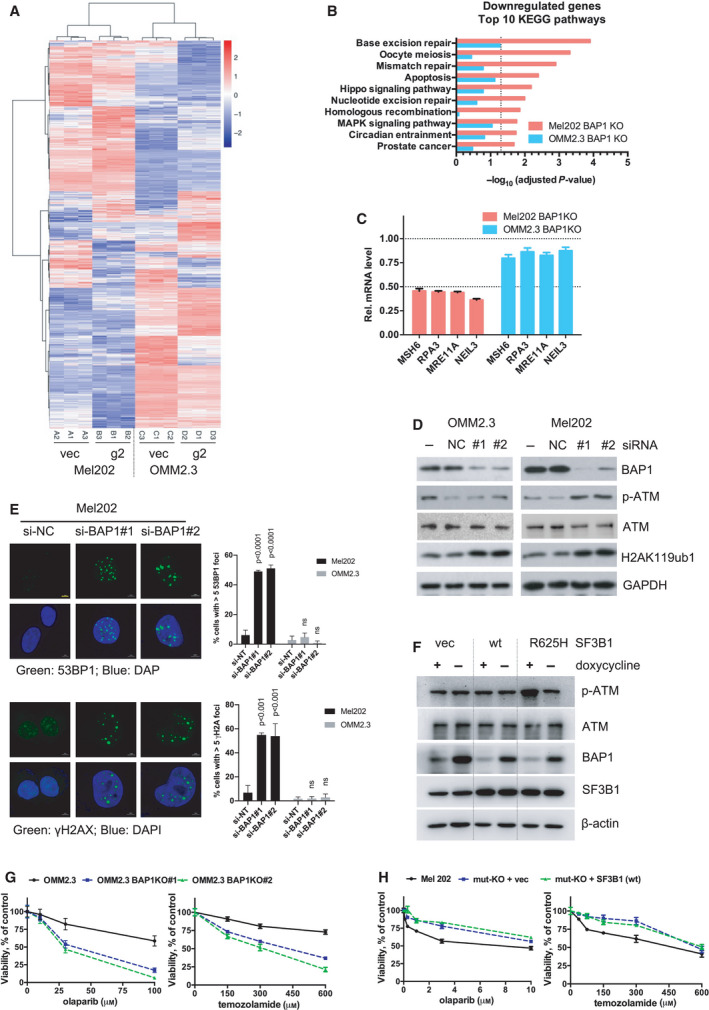
Fig. 4
BAP1 deficiency combined with SF3B1 hotspot mutation induces DNA damage response. (A) RNA‐Seq data of differentially expressed genes (DEGs) in Mel202 and OMM2.3 cells with BAP1 KO (g2) compared with vector control (vec). Data were analyzed with DESeq2 (cutoff: adjusted