- Title
-
Aggressiveness and Metastatic Potential of Breast Cancer Cells Co-Cultured with Preadipocytes and Exposed to an Environmental Pollutant Dioxin: An in Vitro and in Vivo Zebrafish Study
- Authors
- Koual, M., Tomkiewicz, C., Guerrera, I.C., Sherr, D., Barouki, R., Coumoul, X.
- Source
- Full text @ Environ. Health Perspect.
|
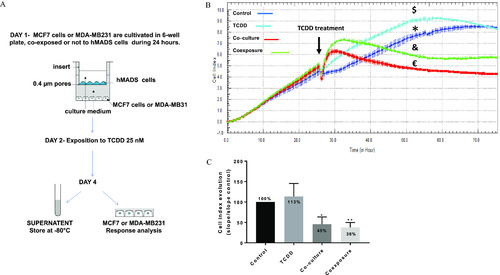
Co-culture model and real time MCF7 cells analysis. (A) Presentation of the 2D co-culture system and the protocol. (B) xCELLigence dynamic monitoring of MCF-7 cells, A representative graph from xCELLigence system: cell index (CI) profiles are the |
|
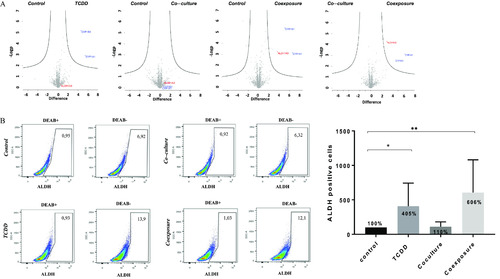
Proteomics analysis and ALDH enzymatic analysis of MCF7 cells growth in co-culture with hMADS and exposed to |
|
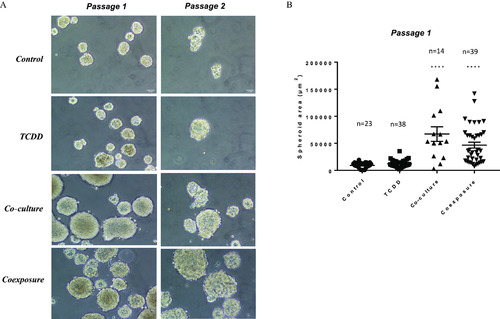
Sphere formation assay. (A) Representative images of sphere formation taken on day 7 on two consecutive generations. Only breast stem/progenitor cells can self-renew and grow into a spheroid structure. Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with 25nM TCDD), co-culture (MCF-7 co-cultured with hMADS) and coexposure (co-culture with TCDD). Scale bar 10μm . (B) Spheroid area (in μm2) in passage 1 using media from the various conditions. Graph represents means±SEM of n (see figure) measurements. The numerical information mean±SEM and p-values are provided in Table S3. Untreated MCF-7 cells alone represent the control condition (Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series, **** p<0.0001 ). Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin. |
|
Morphological differences of MCF7 cells and MDA-MB-231 cells following exposure to TCDD, co-culture, or coexposure. Cells were grown with hMADS and/or treated with |
|
Protein (A-actin, B-beta-catenin) localization & nucleus staining in MCF7 cells and MDA-MB-231 cells. MCF7 cells were grown with hMADS and/or treated with |
|
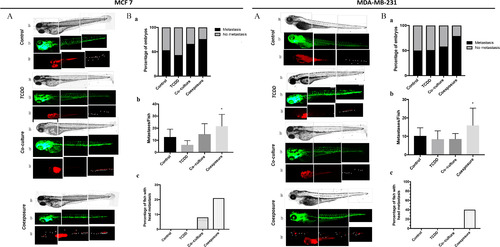
Measurement of metastatic spread of MCF7 and MDA-MB231 cells in zebrafish larvae in vivo models Human RFP-MCF7 (left) or RFP-MDA-MB-231 (right) cells cultured with conditioned media from different conditions, were injected into the perivitelline space of 2-day-old zebrafish larvae [Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with |