- Title
-
Identification of novel susceptibility loci for non-syndromic cleft lip with or without cleft palate
- Authors
- Ma, L., Lou, S., Miao, Z., Yao, S., Yu, X., Kan, S., Zhu, G., Yang, F., Zhang, C., Zhang, W., Wang, M., Wang, L., Pan, Y.
- Source
- Full text @ J. Cell. Mol. Med.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
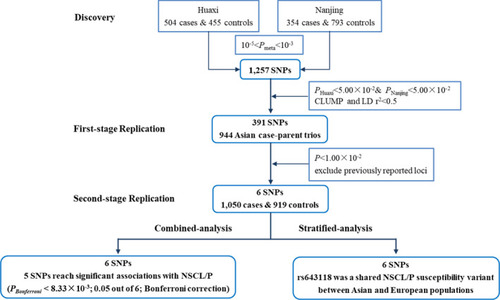
Flow chart of this study |
|
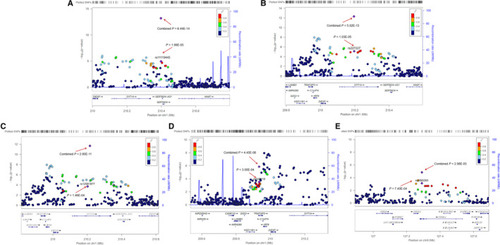
Regional plot of the five newly identified SNPs. The regional plots indicate five variants (A) rs11119445, (B) rs227227, (C) rs12561877, (D) rs643118 and (E) rs2095293 and linkage disequilibrium structure. Logistic regression analyses were used to test the genetic association under an additive model. The marker SNPs are shown as purple diamonds with |
|
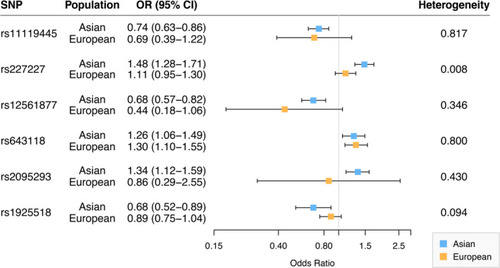
Comparison of associations for the six genetic variants in Asian and European ancestry populations. |
|
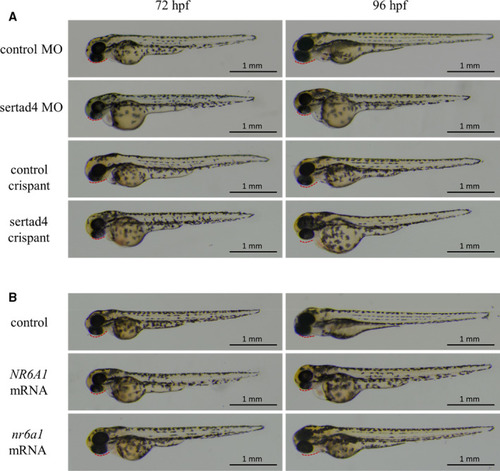
Zebrafish larvae were imaged with transmitted light. (A) Translation‐blocking morpholino and CRISPR/Cas9‐based targeted sertad4 knockdown (crispant) zebrafish models, (B) Embryos with over‐expression of human PHENOTYPE:
|