- Title
-
The role of clonal communication and heterogeneity in breast cancer
- Authors
- Martín-Pardillos, A., Valls Chiva, Á., Bande Vargas, G., Hurtado Blanco, P., Piñeiro Cid, R., Guijarro, P.J., Hümmer, S., Bejar Serrano, E., Rodriguez-Casanova, A., Diaz-Lagares, Á., Castellvi, J., Miravet-Verde, S., Serrano, L., Lluch-Senar, M., Sebastian, V., Bribian, A., López-Mascaraque, L., López-López, R., Ramón Y Cajal, S.
- Source
- Full text @ BMC Cancer
|
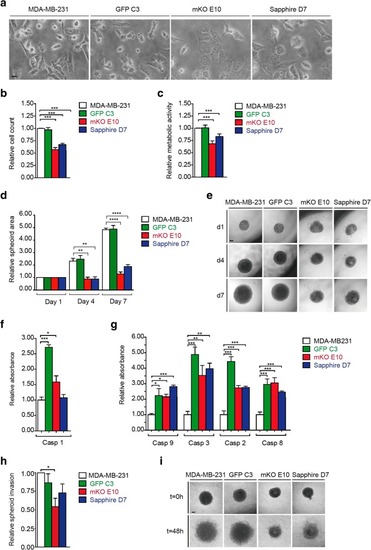
Phenotypic characterization of MDA-MB-231, GFP C3, mKO E10, and Sapphire D7 cell lines. |
|
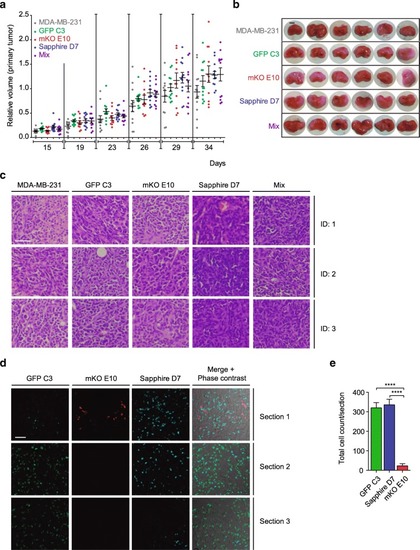
Malignancy of individual clones and tumor heterogeneity. |
|
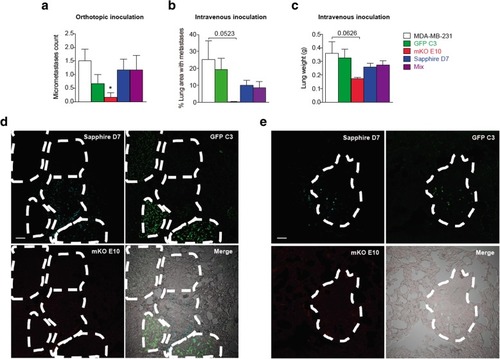
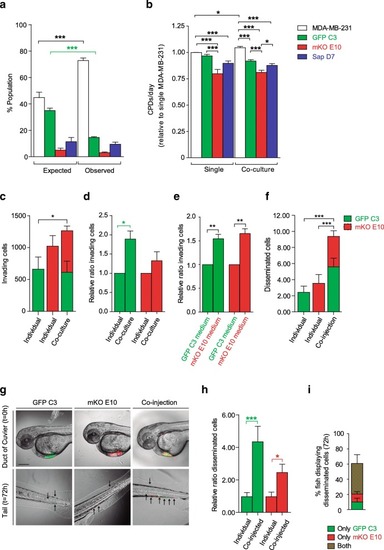
Metastatic capacity of cell lines and an equal mix of all cell lines. |
|
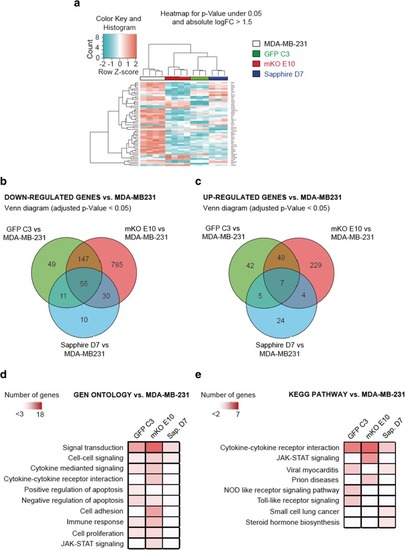
Transcriptional profile of MDA-MB-231 clonal cell lines. |
|
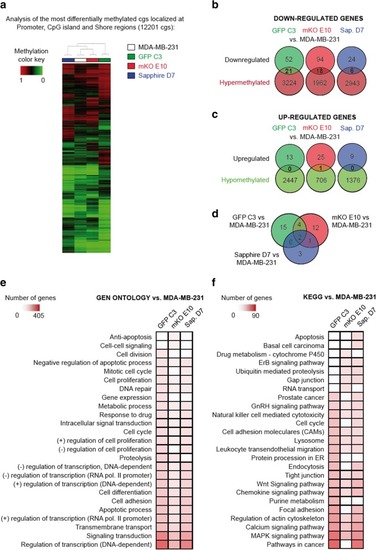
DNA methylation profile of MDA-MB-231 clonal cell lines. |
|
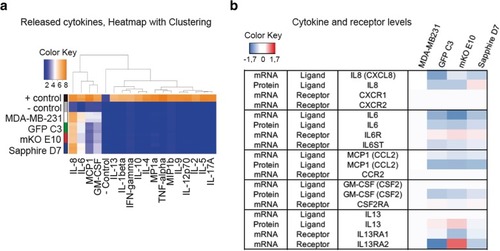
Cytokine expression. |
|
Clone interactions among parental and clonal cell lines in co-culture for 28 days |
|
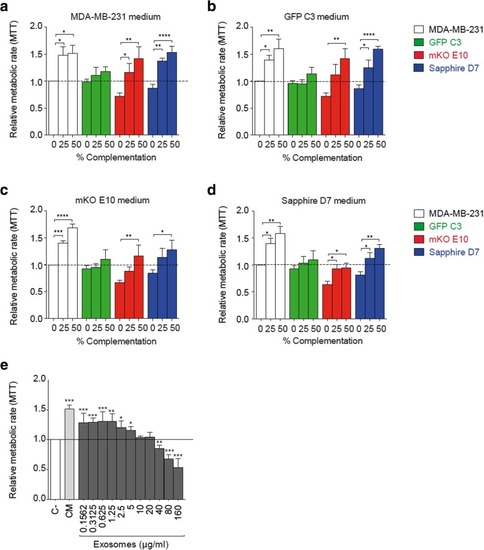
Clone communication by released soluble factors. |