- Title
-
Molecular Evolution of Tryptophan Hydroxylases in Vertebrates: A Comparative Genomic Survey
- Authors
- Xu, J., Li, Y., Lv, Y., Bian, C., You, X., Endoh, D., Teraoka, H., Shi, Q.
- Source
- Full text @ Genes (Basel)
|
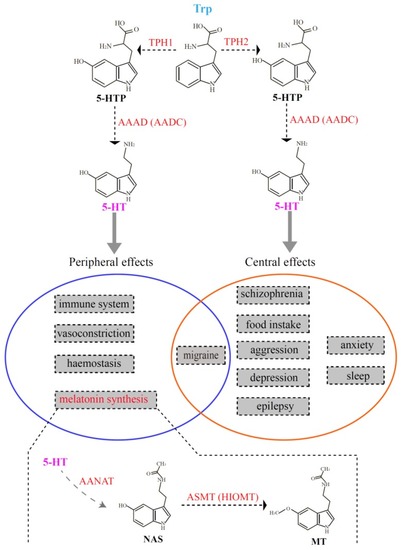
Schematic representation of the two serotonin systems in vertebrates. The top part denotes the process of serotonin biosynthesis, which is also the first two steps in melatonin synthesis. The middle part (in the circles) summarizes the functions regulated by the two serotonin systems. The bottom part interprets the last two steps for melatonin synthesis. 5-HT, 5-hydroxytryptamine; AAAD (AADC), |
|
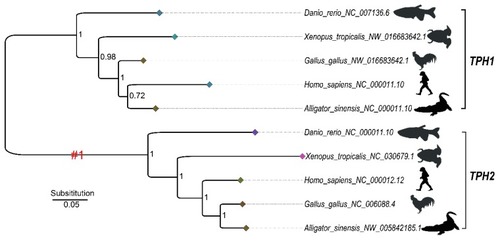
Phylogenetic trees and genome synteny of |
|
Phylogenetic trees and genome synteny of |
|
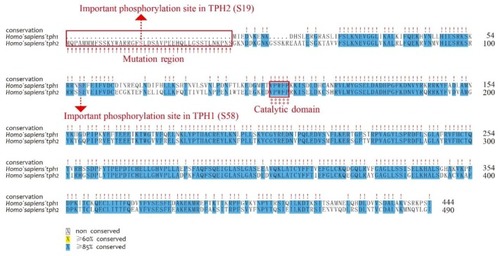
Alignment of human TPH1 and TPH2 protein sequences. Important residues include the mutation region (in the NH2-terminal red box), phosphorylation sites (dashed lines with red arrows), and catalytic domains (‡ with the red box). |
|
Alignment and secondary structures of TPH protein sequences in vertebrates. ( |
|
Alignment and secondary structures of TPH protein sequences in vertebrates. ( |
|
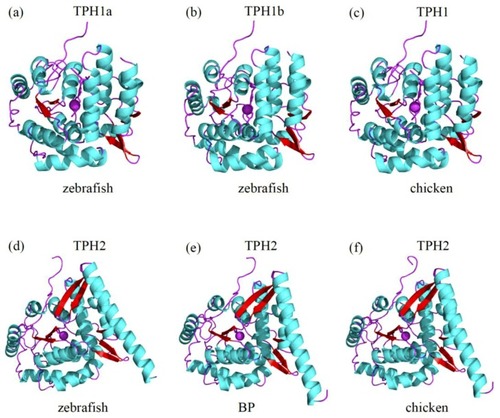
Predicted 3D structures of representative TPH proteins. Comparisons of the 3D structures of zebrafish TPH1a ( |
|
The Bayesian inference (BI) tree based on five TPH1 and TPH2 proteins selected from those shown in |
|
Alignment of the datasets for selection pressure analysis. The five positively selected sites predicted by the branch-site model test are marked with red boxes and emphasized with section marks (§). |