- Title
-
Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility
- Authors
- Goudarzi, M., Berg, K., Pieper, L.M., Schier, A.F.
- Source
- Full text @ Elife
|
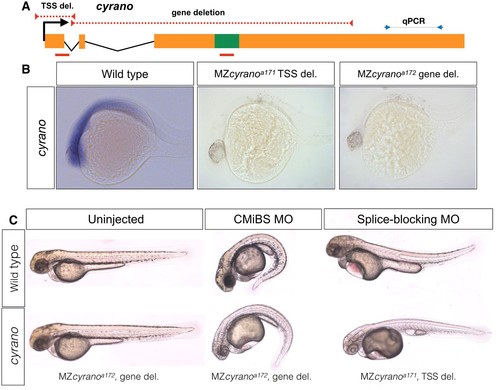
normal embryogenesis of cyrano mutants.(A) The positions of TSS-deletion allele and gene deletion allele are marked by dashed red lines. Green box represents the conserved element in cyrano which is complementary to miR-7. Solid red lines indicate the position of the first exon-intron boundary (e1i1) morpholino and conserved microRNA binding site (CMiBS) morpholinos. Arrows flanking black dotted line mark the primer binding sites for qRT-PCR product. (B) Representative images of in situ hybridization for cyrano in wild type (15/15) and both homozygous TSS-deletion (21/22) and gene deletion (18/18) 1-dpf. (C) At 2-dpf gene deletion mutants (lower-left), (and TSS-deletion mutants, not shown) were not different from the wild-type embryos (upper-left). Morpholino injected wild-type embryos (upper-middle and upper-left) reproduced observed phenotype in Ulitsky et. al (Kok et al., 2015). Morpholino injected deletion-mutants, lacking the corresponding binding sites for morpholinos, (lower-middle and lower-left) were comparable to morpholino injected wild types. PHENOTYPE:
|
|
Normal embryogenesis of gas5 mutants.(A) Position of the TSS-deletion allele in gas5 is marked by dashed red line. Arrows flanking black dotted lines mark the primer binding sites for 5’-qPCR and 3’-qPCR products. (B) Representative in situ hybridization images for gas5 in wild type (11/11) and homozygous TSS-deletion mutants (11/11). (C) Maternal and Zygotic gas5 (MZgas5) mutant embryos at 1-dpf were indistinguishable from the wild-type embryos at the same developmental stage (not shown). (D) Expression level of gas5 and osbpl9 measured by qRT-PCR. Tor3A, the other neighboring gene, was not expressed at the investigated time-point. (E) Expression level of gas5, its trans targets ptena, ptenb and nr3c1measured by qRT-PCR. The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (*, **, ***, and **** respectively mark p-values<0.05,<0.01,<0.001 and<0.0001). |
|
Normal embryogenesis of lnc-setd1ba mutants.(A) The relative position of lnc-setd1ba and the protein-coding gene setd1ba. The gene deletion region is marked by dashed red line. Arrows flanking black dotted line mark the primer-binding sites for qRT-PCR product. (B) Maternal and zygotic lnc-setd1ba mutants were not different from wild-type embryos at 1-dpf. (C) Representative images of in situ hybridization for lnc-setd1ba at four- to eight-cell stage mutant (18/18) and wild-type (25/25) embryos. (D) In situ hybridization for the protein-coding mRNA, setd1ba (9/11) in lnc-setd1ba mutants compared to the wild-type embryos (15/15). (E) qRT-PCR at 1 cell stage and 1-dpf for the lncRNA and its neighboring genes rhoF and setd1ba. The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (ns, *, **, ***, and **** respectively mark p-values≥0.05,<0.05,<0.01,<0.001 and<0.0001). |
|
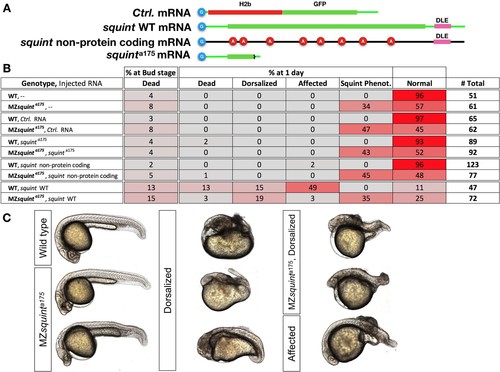
No non-coding function for squint 3’UTR.(A) The position of untranslated regions (brown), coding region (green), putative Dorsal Localization Element- DLE (blue) and the gene deletion (red dashed line) in the squint genomic locus. Arrows flanking black dotted line mark the primer binding sites for qRT-PCR product. (B) In situ hybridization for squint at 8-cell stage on wild-type (18/20) and MZsquinta175(17/17) embryos. (C) qRT-PCR for squint and eif4ebp1 on wild-type and MZsquinta175 embryos at 1-cell stage. (D) Two representative MZsquinta175 embryos. (E) MZsquinta175 embryonic phenotype (N = 4 independent crosses, n = 360 embryos). The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (ns, *, **, ***, and **** respectively mark p-values≥0.05,<0.05,<0.01,<0.001 and<0.0001). PHENOTYPE:
|
|
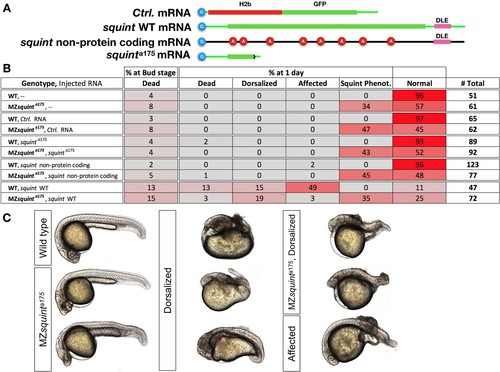
Dorsalization induced by Overexpression of squint mRNA but not its non-protein coding version.(A) Schematic representation of injected mRNAs. Cap-analog is indicated by in blue circles at the beginning of each mRNA. squint non-protein coding mRNA was generated by adding 8 Adenine-nucleotides (red circles) after in-frame ATG codons. (B) Table shows scoring outcome of observed phenotypes in embryos injected with 30 pg of each indicated mRNA. (C) Representative embryos showing typical wild-type, squint mutant or dorsalized morphology. Ambiguous phenotypes were scored as ‘Affected’. |
|
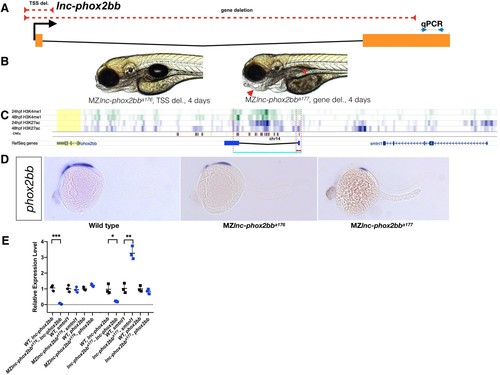
Requirement for lnc-phox2bb genomic elements but not RNA.(A) The red dashed lines depict the respective positions of the lnc-phox2bb TSS and gene deletion. Arrows flanking black dotted line mark the primer binding sites for qRT-PCR product. (B) Homozygous gene deletion mutants but not the TSS-deletion mutants show embryonic defects in jaw formation (arrow head) and swim bladder inflation (asterisk) by 4-dpf. (C) Histone marks (H3K4me1 and H3K27ac) associated with enhancer activity (Bogdanovic et al., 2012) and conserved noncoding elements (CNEs) (Hiller et al., 2013) overlap with gene deletion. (D) phox2bb expression pattern in the TSS and gene deletions. (E) qRT-PCR analysis on MZ TSS-deletion and gene deletion mutants. The statistical significance of the observed changes was determined using t-test analysis and represented by star marks (*, **, ***, and **** respectively mark p-values<0.05,<0.01,<0.001 and<0.0001). |