- Title
-
Acquisition of glial cells missing 2 Enhancers Contributes to a Diversity of Ionocytes in Zebrafish
- Authors
- Shono, T., Kurokawa, D., Miyake, T., and Okabe, M.
- Source
- Full text @ PLoS One
|
Expression of gcm2 in the gills and the cells on the skin surface of zebrafish. Whole-mount in situ hybridization of 5-day-old zebrafish with gcm2 probes. (A) gcm2 is expressed in the gills. (B) Magnification of a yolk sac area. Some cells in the yolk sac express gcm2 (arrowheads). EXPRESSION / LABELING:
|
|
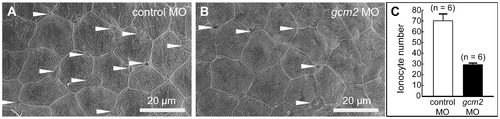
Scanning electron microscopy of the yolk sac membrane in 3-day-old zebrafish. (A, B) Magnification of the yolk sac membrane. (A) An embryo injected with a control morpholino (MO). (B) An embryo injected with a gcm2 MO. (C) Quantitative comparison of the ionocyte number per 0.04 mm2 between embryos injected with control MO and gcm2 MO. The ionocyte number on the yolk sac membrane area in gcm2 morphants is less than that in control MO. Arrowheads indicate ionocytes. Because it is likely that cells were still alive or had differentiated without expression of a marker gene by loss of function, we observed the external morphology of the skin surface in morphants using scanning electron microscopy. PHENOTYPE:
|
|
gcm2 is essential for the development of HRCs and suppresses the development of NCCCs on the skin surface. (A, B) Whole-mount in situ hybridization of zebrafish 48 hours post-fertilization (hpf) with an atp6v1al probe as a HRC marker. (C, D) Whole-mount in situ hybridization with an atp1b1b probe as a NaRC marker. (E, F) Whole-mount in situ hybridization with a slc12a10.2 probe as a NCCC marker. (A, C, E) Zebrafish (48 hpf) injected with a control morpholino (MO). (B, D, F) Zebrafish (48 hpf) injected with a gcm2 MO. (G) Quantitative comparison of atp6v1al-positive cells, atp1b1b-positive cells, and slc12a10.2-positive cells in zebrafish injected with control MO and gcm2 MO. |
|
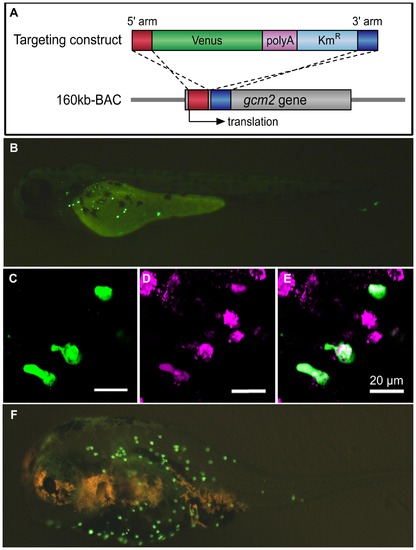
Analysis of gcm2 enhancers specific for ionocytes on the skin surface of zebrafish. (A) Construction of the 160 kb BAC-Venus. The targeting construct was amplified by PCR from a plasmid containing Venus, polyA, and Kmr. Each primer contained 50 bp of the gcm2-derived sequences that served as homology arms for homologous recombination. The 160 kb BAC contained sequences that were 120 kb upstream and 40 kb downstream of the gcm2 locus. After homologous recombination, Venus was inserted into the translation site (160 kb BAC-Venus). (B) A 160 kb BAC-Venus transient transgenic zebrafish (72 hpf). Venus-expressing cells are observed on the skin surface but not in the gills. (C) Several cells on the yolk sac expressed Venus. (D) Ionocytes stained with MitoTracker. (E) Merge of (C) and (D), showing overlap in staining with a subset of ionocytes. (F) A 160 kb BAC-Venus transient transgenic 7-day-old fugu. Venus-expressing cells are observed on the skin surface. |
|
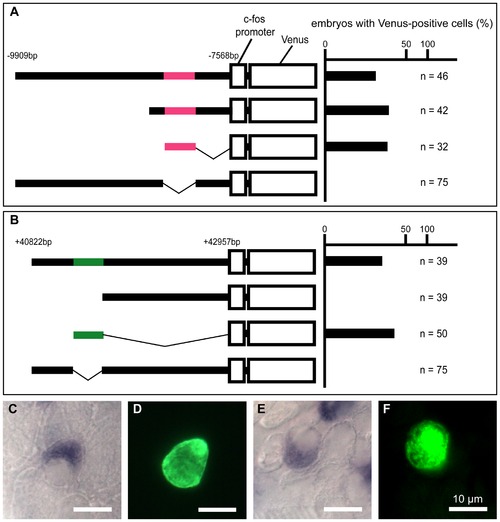
Transient transgenic analysis of the gcm2 enhancer regions for HRCs specific to the skin surface. (A, B) The chart on the right shows the percentage of embryos with Venus-positive cells on the skin surface at 24 hpf. (C–F) In situ hybridization with an atp6v1al probe in combination with immunohistochemistry using anti-GFP on the yolk sac of transient transgenic 4-day-old zebrafish. (C, D) A zebrafish expressing pTolfV that contains the gcm2 enhancer region that is 8 kb upstream. (E, F) A zebrafish expressing pTolfV that contains the gcm2 enhancer region that is 41 kb downstream. The gcm2 loci at -8249 to -8004 bp upstream and at +41277 to +41497 bp downstream were required for Venus expression in HRCs. |