- Title
-
Transcriptomic Profiling Revealed Signaling Pathways Associated with the Spawning of Female Zebrafish under Cold Stress
- Authors
- Ge, G., Long, Y., Song, G., Li, Q., Cui, Z., Yan, H.
- Source
- Full text @ Int. J. Mol. Sci.
|
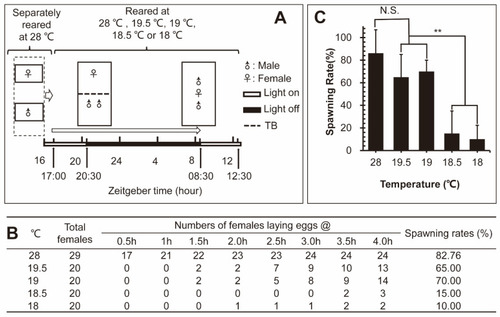
The fecundity of the zebrafish at different temperatures. ( |
|
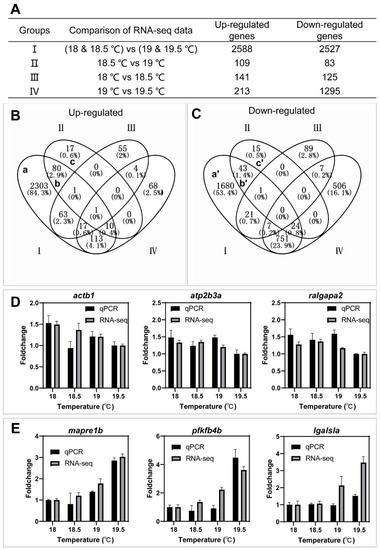
The workflow of RNA−seq data analysis. ( |
|
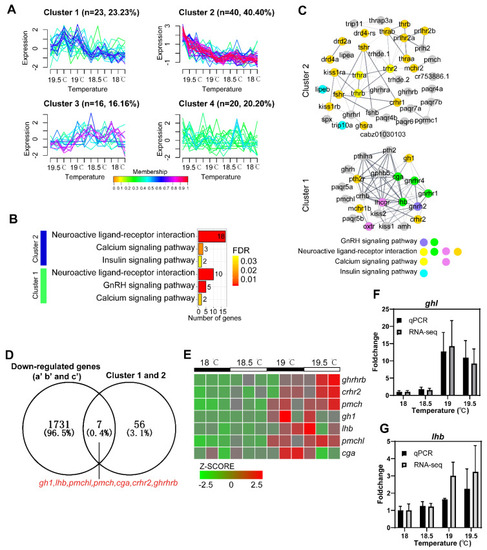
Identification of key genes associated with the spawning of females using Venn diagram. ( |
|
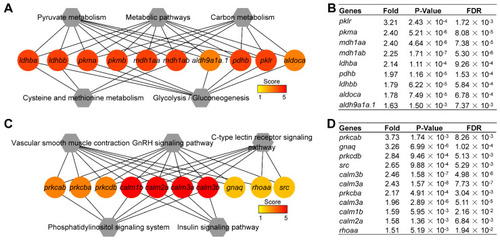
Hub signaling pathways from KEGG enrichment analysis of up- and |
|
Hub genes within the top five hub pathways. ( |
|
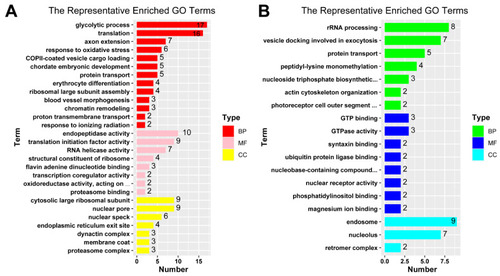
Bar plots for representative terms of GO enrichment analysis. GO terms redundancy were reduced by REVIGO tool to give a representative subset of terms for up-regulated key genes ( |
|
Details of hormone−related genes detected in the female zebrafish brains by RNA−seq. ( |