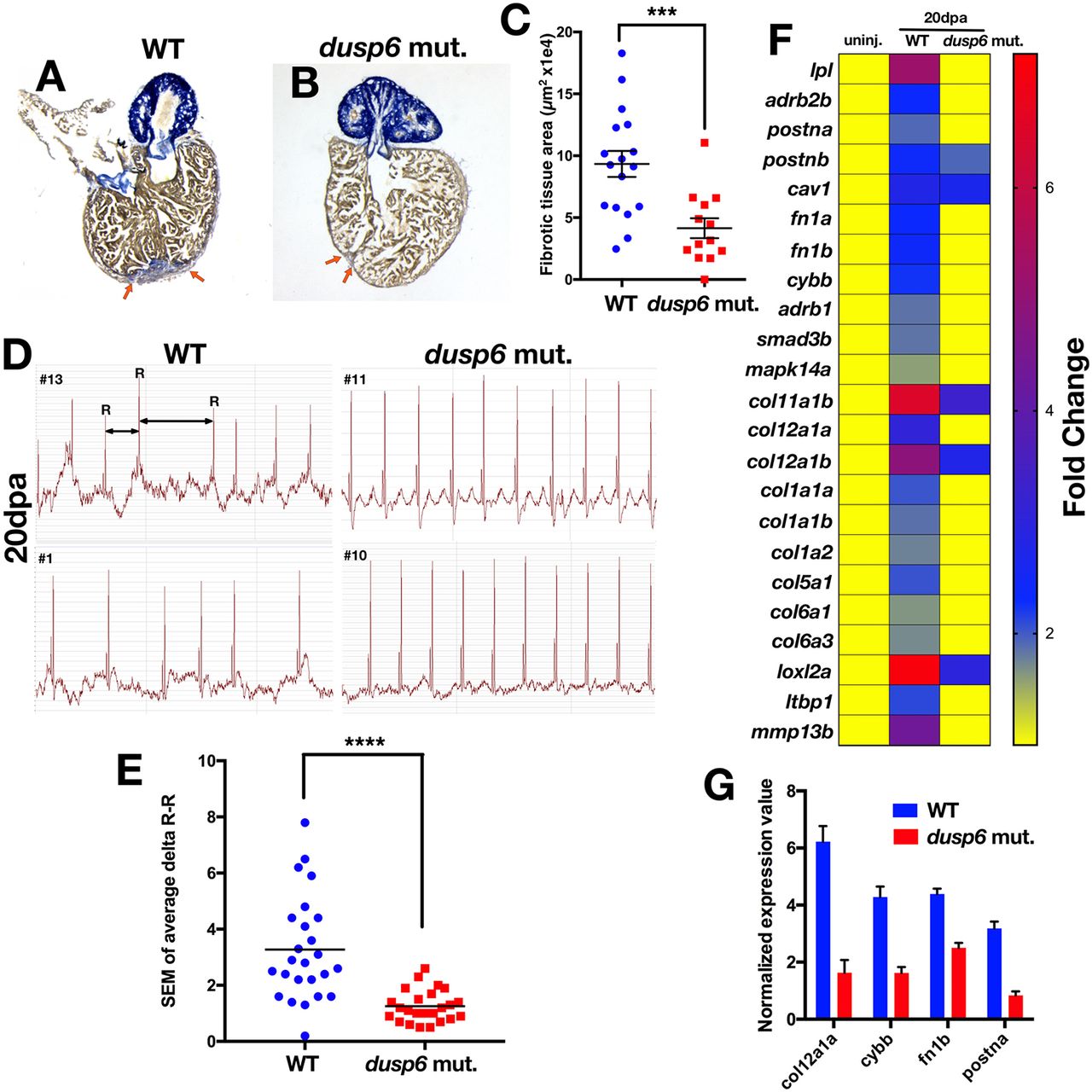
Fig. 4
dusp6 mutant hearts show accelerated regeneration. (A,B) Sections of hearts at 20 dpa stained with AFOG to visualize the deposition of fibrotic tissue (arrows). dusp6 mutant hearts (n=13) (B) have reduced fibrotic tissue area compared with WT hearts (n=17) (A). (C) Quantitation of fibrotic tissue area at 20 dpa. ***P<0.001, Student's t-test. (D) Representative ECG from WT (n=25) and dusp6 mutants (n=24) at 20 dpa. dusp6 mutant fish exhibit fewer arrhythmic events than WT. ECG are shown from four fish (#1, #10, #11, #13). (E) Quantitation of the arrhythmic events, measured as R-R intervals, in WT and dusp6pt30a/pt30a zebrafish at 20 dpa. ****P<0.0001, Student's t-test. (F) RNA-seq analysis of uninjured hearts and hearts at 20 dpa. At 20 dpa, WT hearts express fibrosis genes, but in dusp6pt30a hearts fibrosis genes are downregulated, suggesting faster regeneration. (G) Q-PCR analysis of selected fibrosis genes in hearts at 20 dpa. dusp6pt30a hearts show downregulation of fibrosis genes. Values are normalized to β-actin and rnap expression. Data are from one representative experiment from three independent biological replicates.