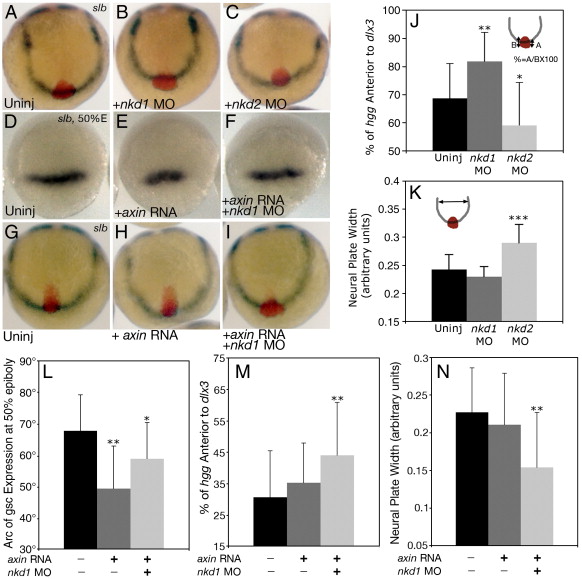
Fig. 8 Reduction of Nkd1 can suppress the axial C&E defect in slb. (A) Uninjected slb mutant. (B) slb embryo injected with nkd1 MO results in a suppression of the axial C&E defect observed in uninjected slb embryos, which is quantified in (J). Note that the convergence of the neural plate remains unchanged (K). In contrast, injection of nkd2 MO exacerbates both the axial C&E (C, J) and the neural plate convergence phenotype in slb mutants (C, K). (D–F) Dorsal view, animal pole to the top of gsc expression at 50% epiboly in slb mutants which are uninjected (D), injected with axin RNA (E) or axin RNA and nkd1 MO (F). The arc of gsc expression is quantified in panel L. (G–I) slb embryos from the same clutch as (D–F) analyzed for percent of axial extension. Anterior view, dorsal to the top of uninjected (G), axin RNA injected (H) and axin RNA and nkd1 MO injected embryos (I). The percentage of axial extension is quantified in panel M and the width of the neural plate is quantified in panel N. (A–C, G–I) 2–3 somite stage. The percentage of complete axial extension is calculated by measuring the proportion of total hgg expression that is over and past the dlx3 expression domain. One hundred percent would be defined as complete extension, as observed, for example, in wild-type (Fig. 7D). Markers are as described in Fig. 7. Error bars represent standard deviation (Students t-test, *, p < 0.05; **, p < 0.01; ***, p < 0.0001).
Reprinted from Developmental Biology, 309(2), Van Raay, T.J., Coffey, R.J., and Solnica-Krezel, L., Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling, 151-168, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.