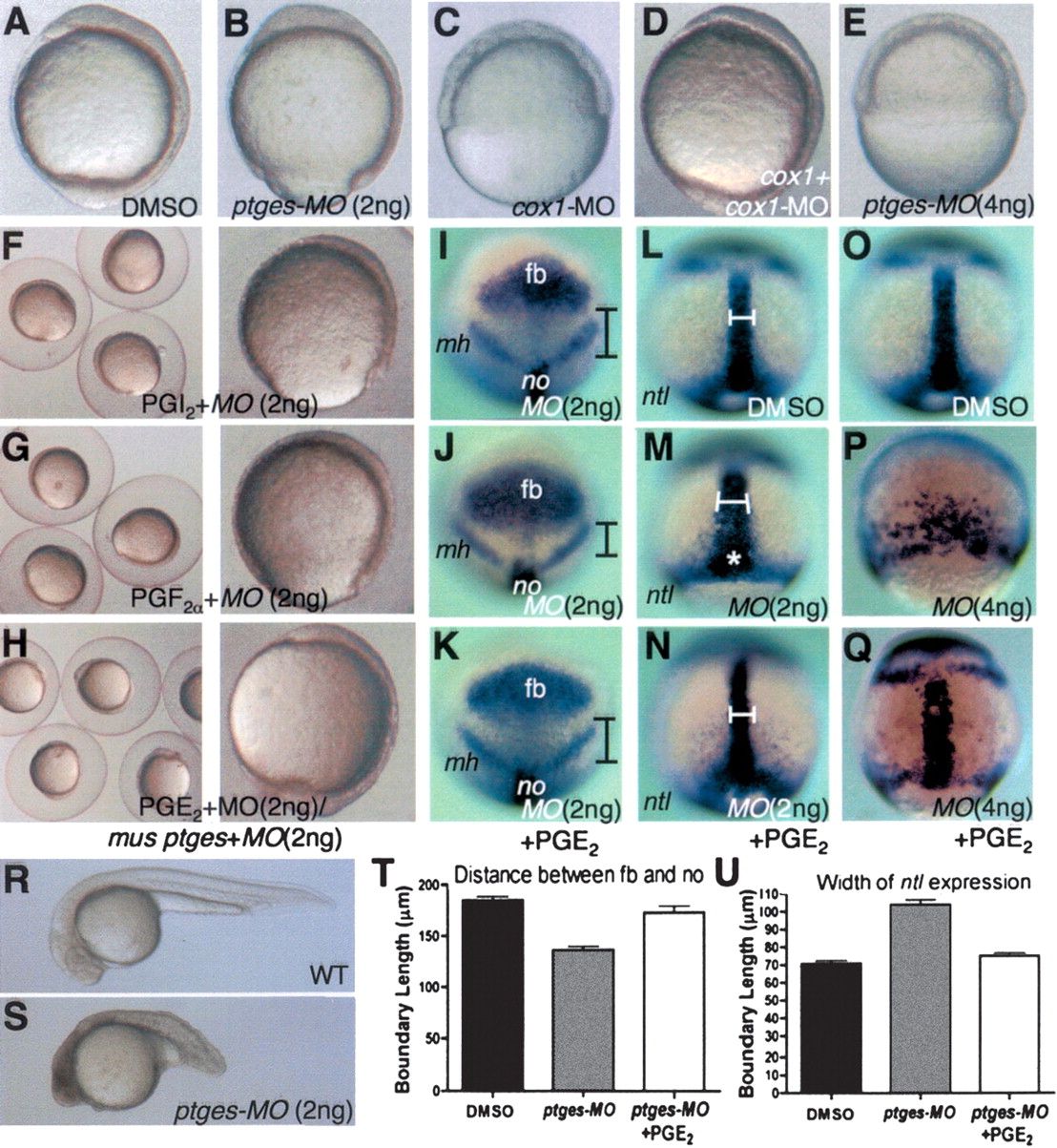
Fig. 2 Ptges-deficient embryos exhibit gastrulation defects that are suppressed by coincubation with PGE2. (A-H) Lateral view, dorsal to the right. (I-K) Animal view, dorsal to the bottom. (L-Q) Dorsal view, anterior to the top. (R,S) Lateral view, anterior to the left. (I-K) Boundaries indicate the distance from the posterior end of the forebrain to the anterior end of the notochord. (L-Q) Boundaries indicate the width of the notochord. (A,I,L,O) Control embryos treated with DMSO. (B,F-H,J,K,M,N,S) Embryos injected with 2 ng of ptges-MO2. (E,P,Q) Embryos injected with 4 ng of ptges-MO2. (C,D) Embryos injected with 10 ng of cox1-MO. (D) Embryos coinjected with cox1-MO (10 ng) and cox1 RNA (50 pg). (F) Embryos injected with 2 ng of ptges-MO2 + PGI2. (G) Embryos injected with 2 ng of ptges-MO2 + PGF2α. (H,K,N) Embryos injected with 2 ng of ptges-MO2 + PGE2. (Q) Embryos injected with 4 ng of ptges-MO2 + PGE2. (H) Embryos coinjected with ptges-MO2 (2 ng) + mouse ptges RNA (100 pg). (I-K) Expression patterns of six3b, pax2.1. (L-Q) Expression pattern of no tail. (T) Quantitative analyses of the distance between the forebrain (fb) and notochord (no) in DMSO, ptges-MO2, and ptges-MO2 + PGE2 (n = 10 per each group). (U) Quantitative analyses of width of ntl expression in DMSO, ptges-MO2, and ptges-MO2 + PGE2 (n = 10 per each group). (fb) Forebrain; (mh) mid-hindbrain; (no) notochord. The white star marks the blastopore to illustrate delayed epiboly. For details see text.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Genes & Dev.