Fig. 3
- ID
- ZDB-FIG-230312-31
- Publication
- Nam et al., 2022 - The Novel Human HtrA2 Ortholog in Zebrafish: New Molecular Insight and Challenges into the Imbalance of Homeostasis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
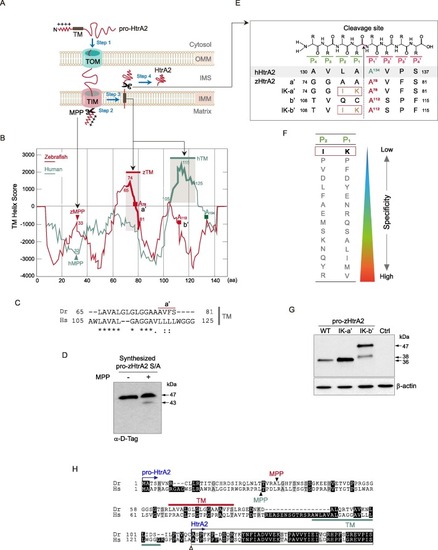
Fig. 3. zHtrA2 is produced via the cleavage of the peptide bond between K111 (P1) and A112 (P1′). (A) Schematic representation of the targeting of HtrA2 to the mitochondrial intermembrane space (IMS). Step 1, Pro-HtrA2 is transported to the mitochondria by the translocase of the outer and inner membranes (TOM and TIM). Step 2, the mitochondrial targeting sequence (MTS) is proteolytically removed by mitochondrial processing peptidase (MPP). Step 3, the transmembrane (TM) domain is inserted laterally into the inner mitochondrial membrane (IMM), where it functions as a hydrophobic anchor. Step 4, HtrA2 is released into the IMS after the proteolytic cleavage on its intermembrane side. (B) The in silico prediction of the putative processing regions in pro-zHtrA2. The TM helix score of pro-HtrA2 was calculated by the TMpred program (prediction of membrane-spanning regions and orientation). The candidate MPP cleavage site (peptide bond between A32 and L33) and the TM segment (AA 65–81) of pro-zHtrA2 were predicted by PSORT II. Red squares indicate the predicted P1′ residues for maturation into zHtrA2 (see Fig. 3E). (C) Conservation of the TM between both pro-HtrA2 proteins. (D) Verification of the proteolytical removal of the MTS by MPP (Step 2 in Fig. 3a). Pro-zHtrA2 S/A was synthesized using the in vitro TNT SP6 protein expression system, incubated with the MPPα/β dimer, and probed with D-Tag Ab. (E) Summary of putative cleavage sites and their mutations in zHtrA2. The cleavage occurs the peptide bond between the P1 and P1′ subsites. Based on A134 at the P1′ subsite of hHtrA2, the A78 (a′) and A112 (b′) in zHtrA2 were predicted to be the P1′ position IK-a′ and IK-b′ denoting the mutants with isoleucine (I) and lysine (K) substitutions at the indicated sites. (F) Illustration of residues with the weakest substrate specificity for the P1 and P2 subsites(Martins et al., 2003). (G) Determination of the cleavage site for the maturation of zHtrA2. The plasmids encoding the indicated pro-zHtrA2 proteins were transfected into HEK293T cells, and the control (Ctrl) cells were transfected with pcDNA3. The proteolytic processed zHtrA2 was analyzed by IB with D-Tag Ab. (H) Summary of sequence alignment of the TM and processing sites for both pro-HtrA2 proteins. Closed and open arrowheads denote the cleavage sites by MPP and for maturation to produce HtrA2, respectively. zHtrA2 starts at residue A112 (see Fig. 3E). |
Reprinted from Gene, 819, Nam, M.K., Moon, J.M., Kim, G.Y., Min Kim, S., Rhim, H., The Novel Human HtrA2 Ortholog in Zebrafish: New Molecular Insight and Challenges into the Imbalance of Homeostasis, 146263, Copyright (2022) with permission from Elsevier. Full text @ Gene