Fig. 4
- ID
- ZDB-FIG-220131-82
- Publication
- Lee et al., 2022 - Host tp53 mutation induces gut dysbiosis eliciting inflammation through disturbed sialic acid metabolism
- Other Figures
- All Figure Page
- Back to All Figure Page
|
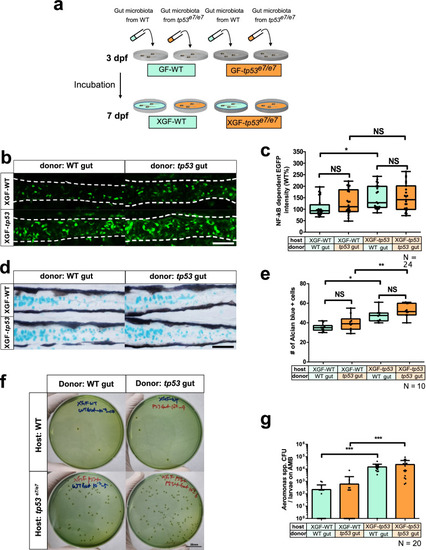
tp53 mutation of the host, not the intestinal bacteria, plays a deterministic role in elevated NF-κB signaling and increased goblet cell numbers. a A schematic for ex-GF experiments where GF-grown WT or tp53 mutant embryos were associated with 7 dpf microbiota of CR WT or tp53 mutants at 3 dpf and examined for their responses at 7 dpf. Final density of microbiota was approximately 104 CFU/mL, as assayed by aerobic growth on TSA plates at 37 °C. b Representative confocal images of the Tg(NFκB:EGFP)-based NF-κB signaling of ex-GF experiments. White dashed lines denote boundaries of GITs based on DIC bright field images. c A boxed plot of NFκB-dependent EGFP fluorescence intensity in ex-GF experiments. Significant differences were identified only in ex-GF tp53 mutant host associated with WT microbiota. N = 24 each. d Representative images of Alcian blue-positive goblet cells of ex-GF experiments. e A boxed plot of the Alcian blue-positive goblet cell number in ex-GF experiments. N = 10 each. f Representative pictures of bacterial cultures from ex-GF experiments using Aeromonas medium base (AMB) plates. Aeromonas spp. was identified based on manufactural criteria (refer to Methods for details). Scale bar = 20 mm. g A bar graph showing a comparison of the Aeromonas spp. CFUs of the ex-GF experiment in AMB plates. The number of Aeromonas spp. colonies in tp53 mutants host associated with WT or tp53 mutant microbiota dramatically increased, whereas those of WT host did not. N = 20 each. The boxed plots and the bar graph were statistically estimated by non-parametric Friedman test followed by Dunn’s multiple comparisons test. Data are represented as mean ± SEM. *< p = .05; **< p = 0.01; ***< p = 0.005. NS not significant |