Figure 5
- ID
- ZDB-FIG-201130-71
- Publication
- Küssau et al., 2020 - Functional Characterization of the N-Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus
- Other Figures
- All Figure Page
- Back to All Figure Page
|
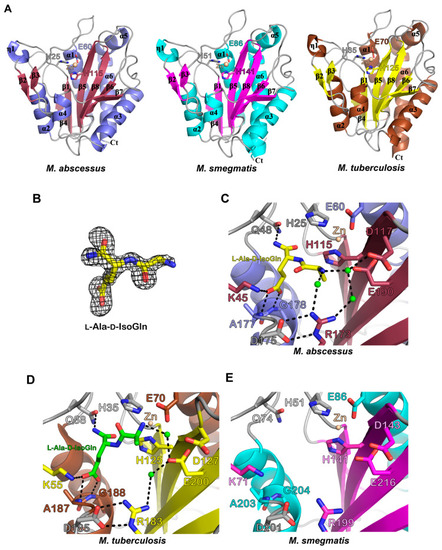
Structural comparisons of the Ami1Mab, Ami1Mtb, and Ami1Msm. ( |