Fig. 3
|
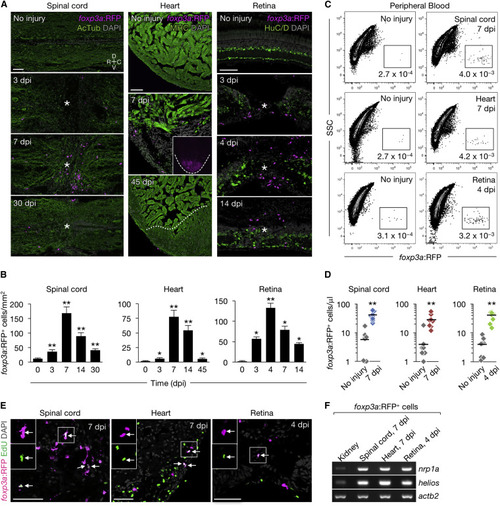
Zebrafish foxp3a+ Cells Infiltrate Regenerating Tissues (A) Sections of injured and uninjured foxp3a:RFP tissues. Immunofluorescence for acetylated Tubulin (AcTub), myosin heavy chain(MHC), and HuC/D marks nerves, cardiac muscle, and neuronal cells, respectively. Asterisks indicate injury epicenter. The dashed line in the inset outlines the ventricle, and the dotted line indicates the border of the regenerated muscle. (B) Quantification of foxp3a:RFP+ cells in A (mean ± SEM, n = 5–8). Uninjured tissues are indicated as 0 dpi. (C) FACS analysis of peripheral blood foxp3a:RFP+ cells. The percentages of cells in each fraction are shown in the plot. (D) Quantification of foxp3a:RFP+ cells in (C) (n = 8). (E) EdU labeling of injured foxp3a:RFP tissues. EdU was injected intraperitoneally at 4, 5, and 6 days after spinal cord and heart injury, or 3 days after retina injury. Arrows point to co-labeled cells. Insets: single-channel images of the marked regions. (F) Semi-qRT-PCR analysis of purified foxp3a:RFP+ cells. C, caudal; D, dorsal; R, rostral; V, ventral. Confocal projections of z stacks are shown in (A) and (E). ∗p < 0.01, ∗∗p < 0.01, Mann-Whitney U test. Scale bars, 50 μm. |
| Genes: | |
|---|---|
| Antibodies: | |
| Fish: | |
| Conditions: | |
| Anatomical Terms: | |
| Stage: | Adult |
| Fish: | |
|---|---|
| Conditions: | |
| Observed In: | |
| Stage: | Adult |
Reprinted from Developmental Cell, 43, Hui, S.P., Sheng, D.Z., Sugimoto, K., Gonzalez-Rajal, A., Nakagawa, S., Hesselson, D., Kikuchi, K., Zebrafish Regulatory T Cells Mediate Organ-Specific Regenerative Programs, 659-672.e5, Copyright (2017) with permission from Elsevier. Full text @ Dev. Cell