Fig. S5
- ID
- ZDB-FIG-141007-84
- Publication
- Pillai-Kastoori et al., 2014 - Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
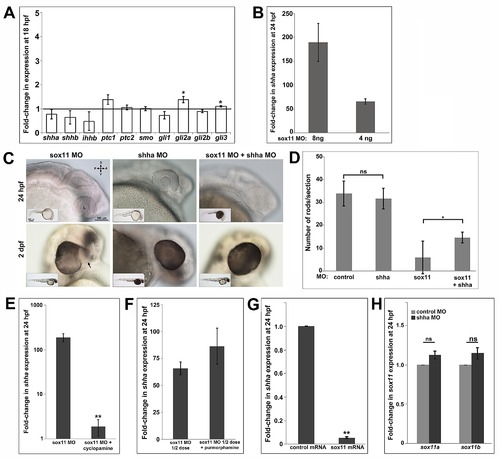
Hh pathway gene expression changes in sox11 morphants. (A) QPCR performed on mRNA from sox11 morphant and control heads at 18 hpf reveal small increases in gli2a and gli3 expression in sox11 morphants compared to controls, but no significant change in shha expression. Relative transcript abundance was normalized to atp5h levels and is presented as the mean fold-change in expression relative to controls (B) At 24 hpf, sox11 morphants demonstrated a large increase in shha expression, which correlated with the dose of sox11 MO injected. Relative transcript abundance was normalized to gapdh levels and is presented as the mean fold-change in expression relative to controls (n = 60 embryos per group, 3 independent biological repeats) *p<0.01, Student′s t-test. (C) Representative bright-field images of embryos injected with sox11 MO alone (left side), shha MO alone (middle), or both shha and sox11 MOs (right side), taken from the set of embryos analyzed in Figure 6E. (D) Co-knockdown of shha increased rod photoreceptor number in sox11 morphants at 3 dpf (number of embryos analyzed: control MO, n = 11; shha MO, n = 10; sox11 MO, n = 16, sox11+ shha MO, n = 15, 3 independent repeats) *p<0.05, Student′s t-test. (E) QPCR performed on mRNA from heads of sox11 morphants treated with vehicle (100% ethanol) or cyclopamine and compared to control morphants treated with vehicle revealed a significant reduction in shha expression in sox11 morphants treated with cyclopamine at 24 hpf. Relative transcript abundance was normalized to gapdh levels and is presented as the mean fold-change in expression relative to controls (n = 40 embryos per group, 3 independent biological repeats) **p<0.0001, Student′s t-test. (F) QPCR for shha was performed on mRNA from the 24 hpf heads of sox11 morphants injected with half the normal dose and treated with DMSO, sox11 morphants (half dose) treated with purmorphamine, and compared to control morphants treated with DMSO. An increase in shha expression was detected in sox11 morphants (half dose) treated with purmorphamine compared to sox11 morphants (half dose) treated with DMSO, however the increase did not reach the threshold for statistical significance. Relative transcript abundance was normalized to gapdh levels and is presented as the mean fold-change in expression relative to controls (n = 40 embryos per group, 3 independent biological repeats). (G) QPCR was performed on mRNA from the 24 hpf heads of zebrafish embryos injected with control (td-tomato) mRNA and embryos injected with zebrafish sox11 mRNA. This analysis revealed a significant decrease in shha expression in embryos overexpressing zebrafish sox11 mRNA compared to the controls. Relative transcript abundance was normalized to 18s rRNA levels and is presented as the mean fold-change in expression relative to controls (n = 30 embryos per group, 3 independent biological repeats). **p<0.0001, Student′s t-test. (H) QPCR performed on mRNA from heads of 24 hpf embryos injected with shha MO or control MO revealed no significant change in expression of either sox11a or sox11b in shha morphants compared to controls. Relative transcript abundance was normalized to gapdh levels and is presented as the mean fold-change in expression relative to controls (n = 45 embryos per group, 3 independent biological repeats). ns, p>0.05, Student′s t-test. D, dorsal; V, ventral; A, anterior; P, posterior; MO, morpholino; hpf, hours post fertilization; dpf, days post fertilization; L, lens. |