|
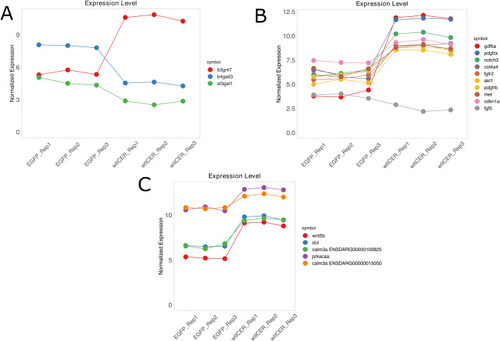
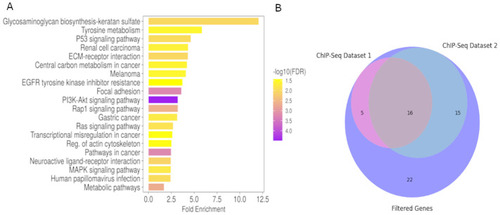
Pathway enrichment. (A) Bar chart of ShinyGO output of all DEG from the zebrafish tumor cell lines for EGFP (n=3) and wtICER (n=3). We note that Glycosaminoglycan biosynthesis of keratan sulfate, while the highest fold-enrichment, PI3K-Akt includes the lowest FDR, and includes many relevant genes involved in melanoma and other cancers including akt1, cdkn1a, col4a4, fgf5, fgfr2, met, pdgfra, pdgfrb. (B) Venn diagram illustrating the overlap of genes identified through analyses of ICER's transcriptional mechanisms. Total filtered list of 58 zebrafish and orthologous Human genes that were upregulated in wtICER, but downregulated in EGFP and phosphorylation mutant, and also contain CRE motifs in the promoter regions; 16 total genes overlap between 2 ChIP-Seq experiments from ENCODE (crema, zcchc24, prkacaa, iqsec1b, rgs12b, sik2b, gli2a, gab1, ttyh3b, gyg1b, zbtb16b, actn4, junba, plxdc2, plpp3, and hmga2).
|