- Title
-
Genome-wide antibiotic-CRISPRi profiling identifies LiaR activation as a strategy to resensitize fluoroquinolone-resistant Streptococcus pneumoniae
- Authors
- Sewgoolam, B., Jim, K.K., de Bakker, V., Bock, F.P., Gibson, P.S., Veening, J.W.
- Source
- Full text @ Nat. Commun.
|
CRISPRi-seq identifies pneumococcal genes influencing fluoroquinolone susceptibility at a genome-wide level. |
|
The ssDNA repair complex RecJFOR is crucial for pneumococcal survival during fluoroquinolone treatment |
|
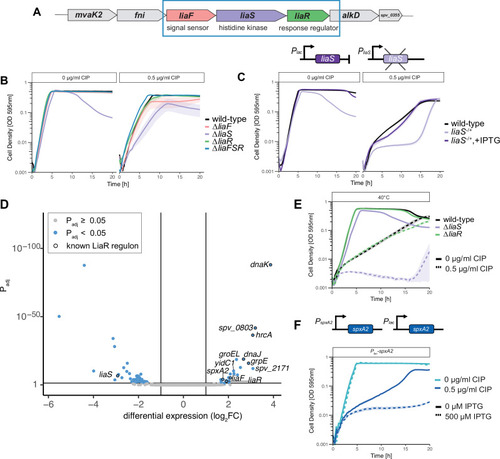
Deleting |
|
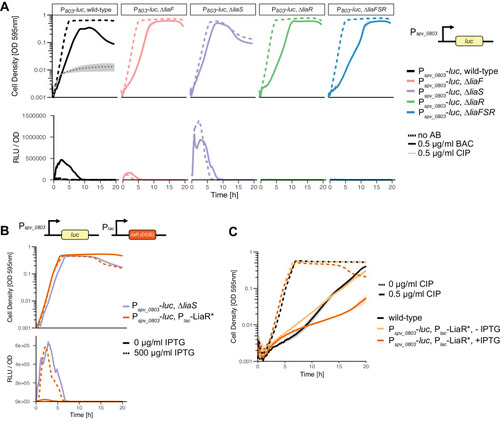
LiaS acts primarily as a phosphatase or inhibitor of LiaR phosphorylation. |
|
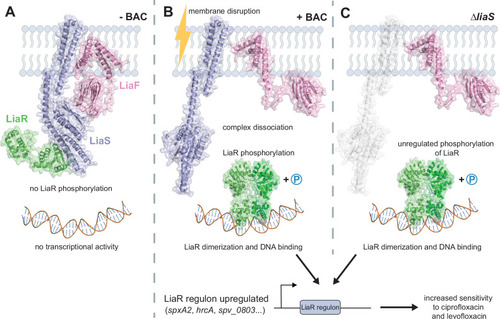
Schematic representation of the role of the LiaFSR three-component regulatory system in fluoroquinolone susceptibility. Hypothetical model of the LiaFSR system modeled with AlphaFold. |
|
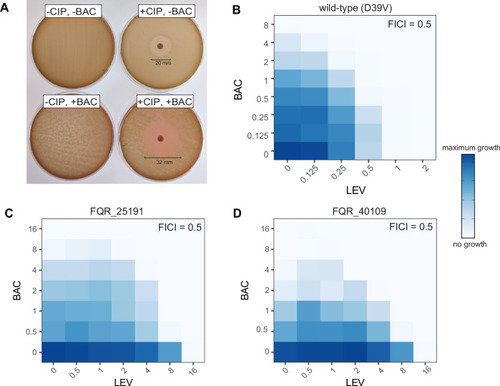
A combination of bacitracin together with ciprofloxacin or levofloxacin increases the potency of these fluoroquinolones. |
|
Levofloxacin and bacitracin act synergistically in vivo in a zebrafish embryo meningitis model. |