- Title
-
miRNAs from Zebrafish Embryo Extracts Inhibit Breast Cancer Invasiveness and Migration by Modulating miR-218-5p/PI3K Pathway
- Authors
- Monti, N., Antinori, D., Proietti, S., Piombarolo, A., Querqui, A., Lentini, G., Liguoro, D., Aventaggiato, M., Lucarelli, M., Pensotti, A., Giuliani, A., Tafani, M., Fuso, A., Bizzarri, M.
- Source
- Full text @ Int. J. Mol. Sci.
|
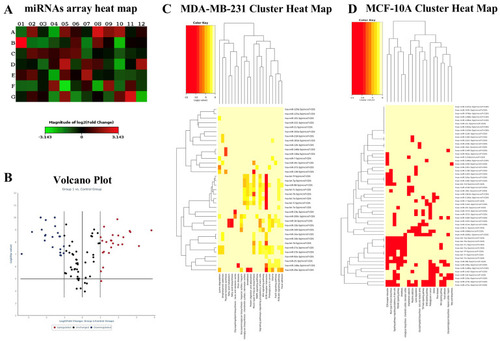
miRNA sequence analysis performed on zebrafish embryos. Data are expressed as raw counts above noise floor. |
|
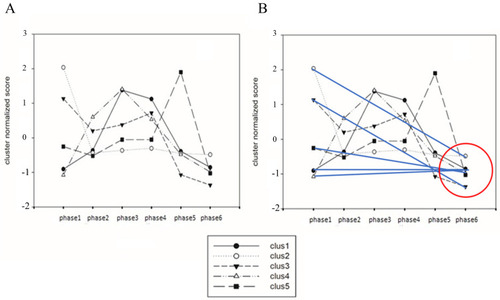
Dynamics of miRNAs are reported ( |
|
|
|
|
|
Array analysis of miRNAs. Heat map ( |
|
|
|
|
|
|
|
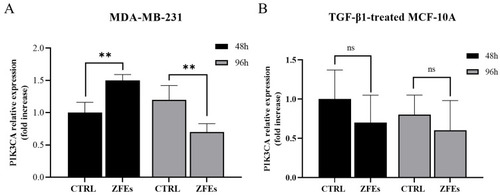
( |
|
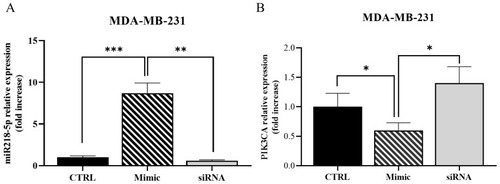
( |
|
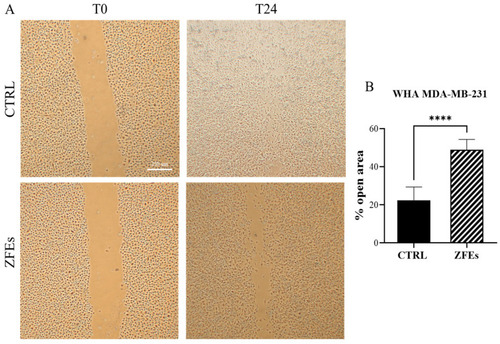
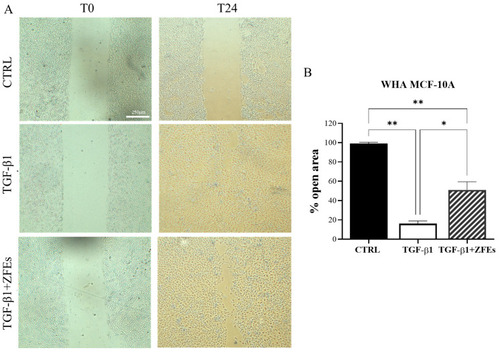
( |
|
|
|
|
|
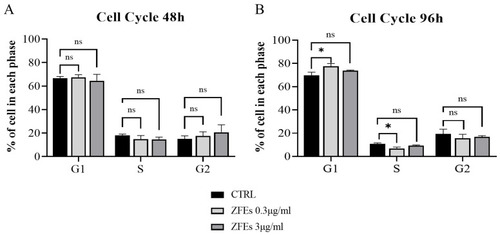
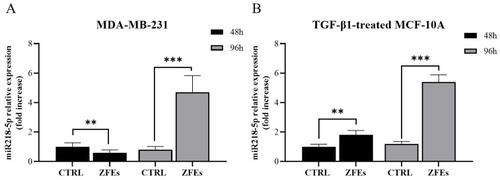
Graphs illustrating the relative expression of p53 ( |