- Title
-
CITK modulates BRCA1 recruitment at DNA double strand breaks sites through HDAC6
- Authors
- Iegiani, G., Pallavicini, G., Pezzotta, A., Brix, A., Ferraro, A., Gai, M., Boda, E., Bielas, S.L., Pistocchi, A., Di Cunto, F.
- Source
- Full text @ Cell Death Dis.
|
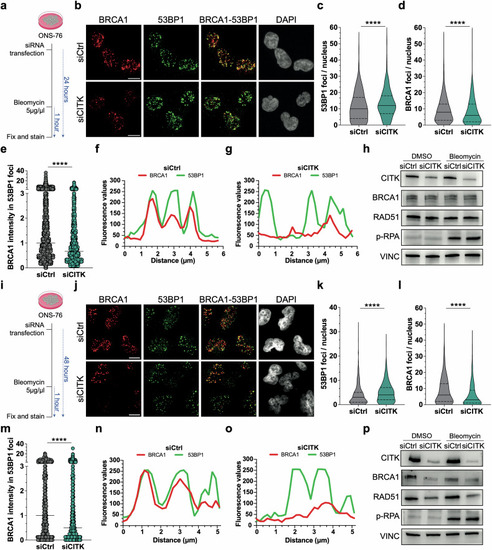
CITK knockdown reduces BRCA1 levels and colocalization between BRCA1 and 53BP1 after induction of DNA damage. |
|
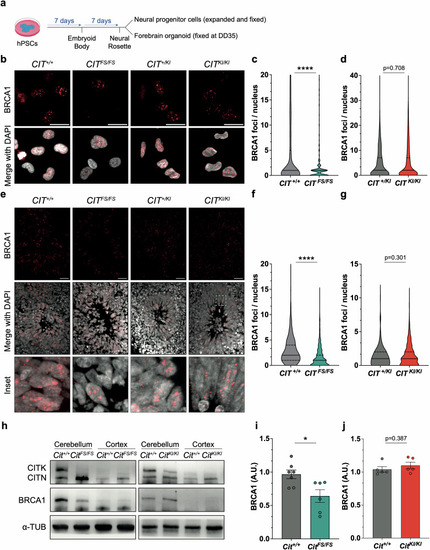
CIT-K knockdown reduces BRCA1 levels independently from proliferative state. |
|
CITK scaffold activity, but not kinase activity, is required for regulation of BRCA1 levels during neurodevelopment. |
|
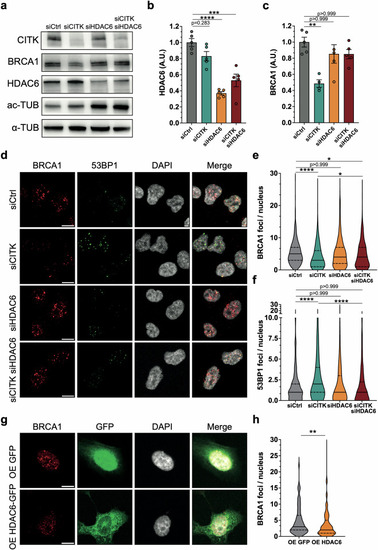
CITK regulates acetylated tubulin levels and co-immunoprecipitates with HDAC6. |
|
HDAC6 targeting recovers BRCA1 levels and DNA damage in CITK depleted cells. |
|
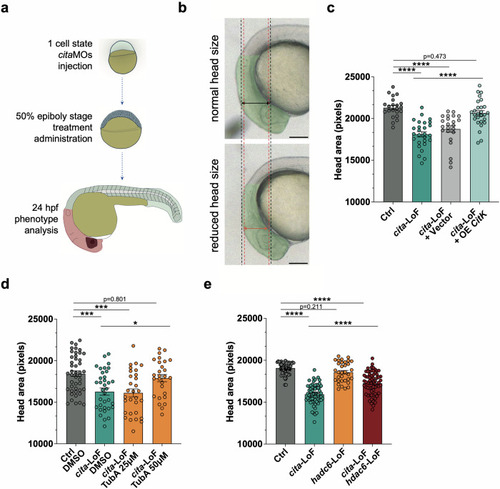
Targeting HDAC6 recovers microcephaly in MCPH17 Zebrafish model. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|