- Title
-
The 419th Aspartic Acid of Neural Membrane Protein Enolase 2 Is a Key Residue Involved in the Axonal Growth of Motor Neurons Mediated by Interaction between Enolase 2 Receptor and Extracellular Pgk1 Ligand
- Authors
- Lee, B.C., Tsai, J.C., Huang, Y.H., Wang, C.C., Lee, H.C., Tsai, H.J.
- Source
- Full text @ Int. J. Mol. Sci.
|
Construction of Eno2 mutant protein expression plasmid. A two-step PCR method was used to generate the expression plasmid, with the mutation of aspartic acid (D) to serine (S) at position 419 (Eno2-[D419S]) as an example. Four primers (p1 to p4) were used for the PCR, where p1 and p4 represent the forward and reverse flanking primers of the |
|
Western blot analysis demonstrated that the recombinant Eno2-wb protein was expressed in the Eno2-knockdown cells transfected with siRNA. |
|
The effect of various point mutations in Eno2 on promoting the neurite growth of motor neurons. ( |
|
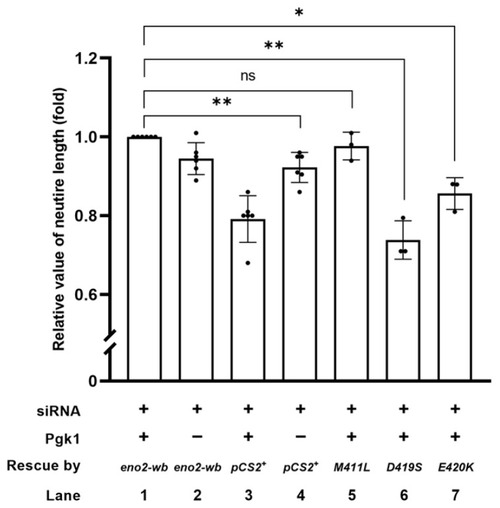
The synergistic effect of promoting motor neurons by the addition of Pgk1 and transfection of various mutations of Eno2. Statistical analysis of the average length of neurites. NSC34 neural cells transfected with siRNA (pCS2+) to inhibit endogenous mouse Eno2 without the addition of extracellular Pgk1 (ePgk1) served as a negative control. As the positive control, NSC34 cells were transfected with |
|
The rescue effect of mutant PHENOTYPE:
|
|
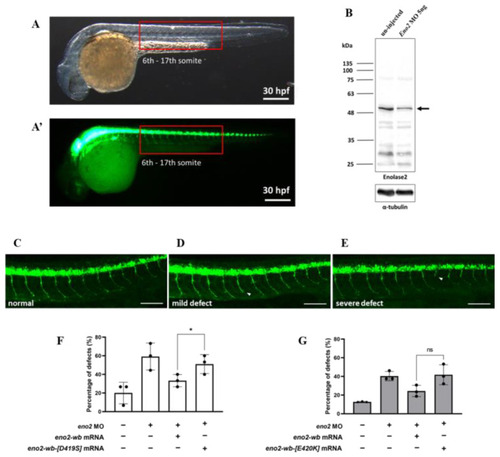
The effect of overexpressing mutant Eno2 on the occurrence rate of branched axons of motor neurons in zebrafish embryos immersed with recombinant Pgk1. ( |
|
Using Western blot to analyze the level of phosphorylated Cofilin (p-Cofilin) expressed in NSC34 cells. ( |
|
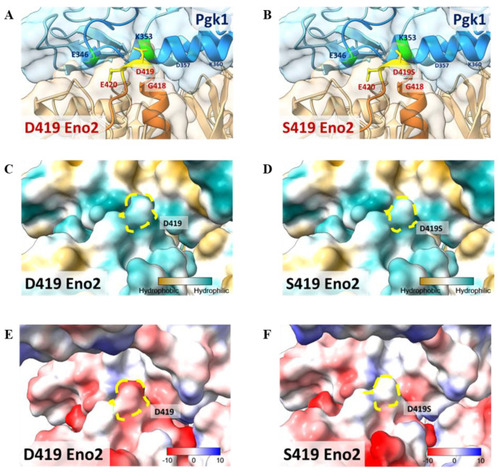
Molecular docking model to illustrate the key amino acid involved in Eno2-ePgk1 interaction. ( |