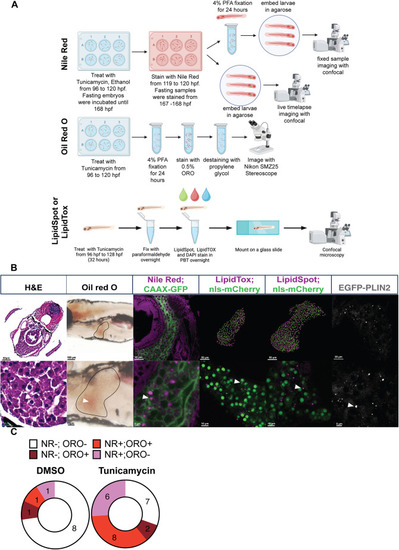
Toolbox to assess hepatic lipid droplets (LDs) in zebrafish larvae. (A) Treatment protocol for inducing steatosis in zebrafish embryos with either tunicamycin (TM) or fasting to 168 h post fertilization (hpf) [7 days post fertilization (dpf)] to assess the use of Oil Red O (ORO), Nile Red (NR), LipidTox and LipidSpot to detect LDs. DAPI, 4′,6-diamidino-2-phenylindole; PBT, phosphate buffered saline containing Tween 20; PFA, paraformaldehyde. (B) Comparison of methods used to detect LD accumulation in the liver of 5 dpf larvae following treatment with TM. Histological [Haematoxylin and Eosin (H&E)] assessment of steatosis compared to dye-based evaluation of LD staining as outlined in A. Staining with ORO, NR, LipidSpot, LipidTox and the transgenic line Tg(fabp10a:EGFP-PLIN2) shows LDs in hepatocytes as viewed by light stereomicroscopy (ORO), multiphoton microscopy (LipidTox and LipidSpot, Imaris rendering of 3D-confocal z-stacks) or confocal microscopy (NR and EGFP-PLIN2, single z-section). LipidTox and LipidSpot samples were imaged along with a transgenic marker for the nucleus Tg(fabp10a:NLS-mCherry), and the NR images were obtained in larvae with EGFP targeted to the hepatocyte cell membrane [Tg(fabp10a:CAAX-EGFP)]. Scale bars: 5, 10, 20, 50 and 100 µm as indicated on each image individually. The images show a typical liver with LD accumulation detected by each staining method as viewed from over 100 embryos from over 20 clutches per staining method. (C) NR is more sensitive than ORO at detecting LD presence in hepatocytes. Larvae were treated with 0.5 µg/ml TM or dimethyl sulfoxide (DMSO) from 96 to 120 hpf, collected and stained with NR, imaged using confocal microscopy, scored as negative or positive for steatosis, then recovered as individual larvae and processed for ORO staining, and scored as negative or positive for steatosis based on the presence of ORO-labelled LDs. Each larva was categorized based on the steatosis score for both dyes. The experiment was carried out on a total of 34 larvae from two clutches.
|