- Title
-
TnP and AHR-CYP1A1 Signaling Crosstalk in an Injury-Induced Zebrafish Inflammation Model
- Authors
- Disner, G.R., Fernandes, T.A.M., Nishiyama-Jr, M.Y., Lima, C., Wincent, E., Lopes-Ferreira, M.
- Source
- Full text @ Pharmaceuticals (Basel)
|
|
|
|
|
|
|
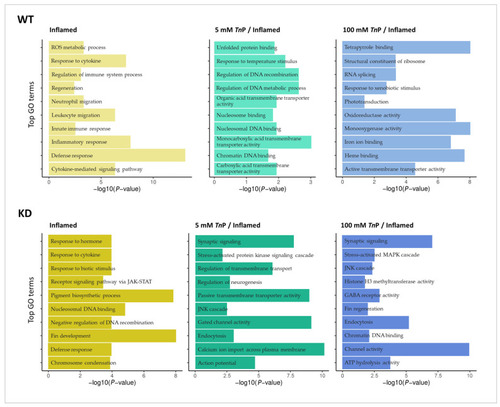
Transcriptomic analysis in a zebrafish inflammation model treated with |
|
Effect of |
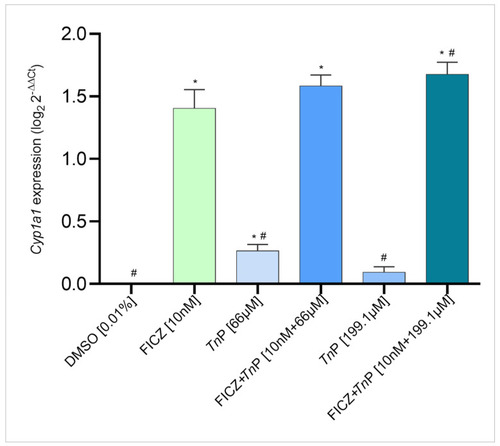
|
Effect of |
|
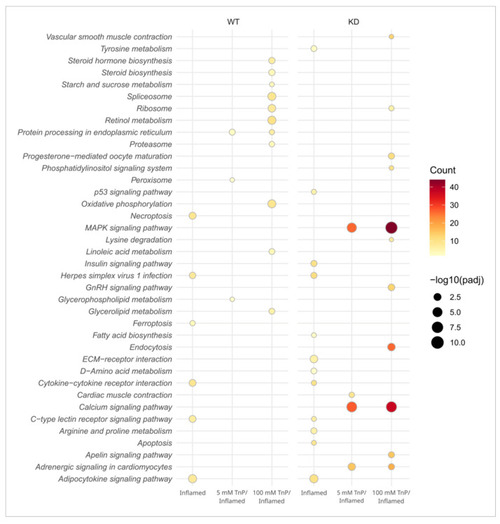
Dot plot illustrating the effect of |
|
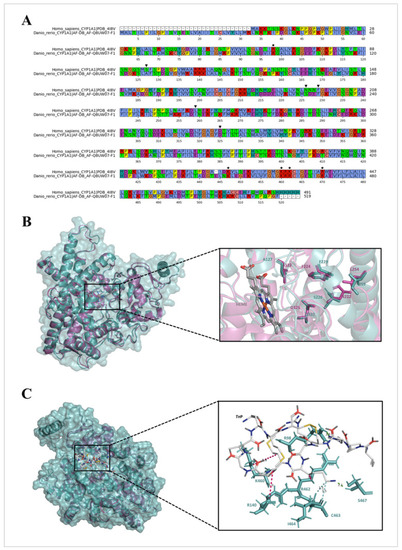
Structure comparison and molecular docking simulations. ( |