- Title
-
Zebrafish Suppressor of Cytokine Signaling 4b (Socs4b) Is Dispensable for Development but May Regulate Epidermal Growth Factor Receptor Signaling
- Authors
- Trengove, M., Rasighaemi, P., Liongue, C., Ward, A.C.
- Source
- Full text @ Biomolecules
|
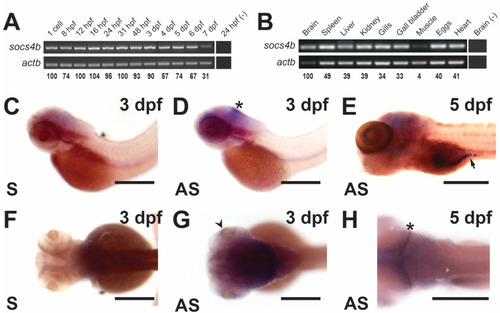
Spatio-temporal expression profile of the zebrafish |
|
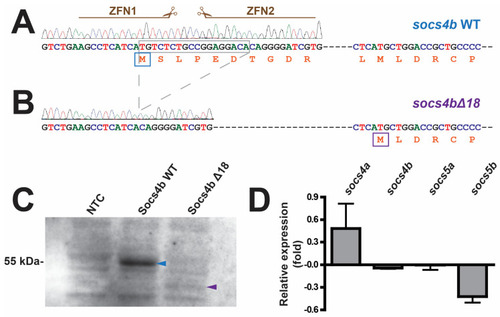
Zinc-finger nuclease-mediated targeting of the zebrafish |
|
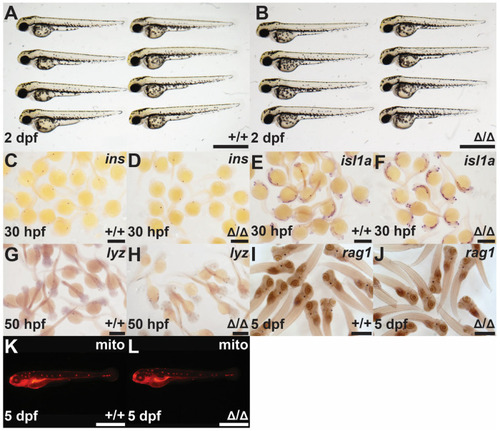
Investigation of the |
|
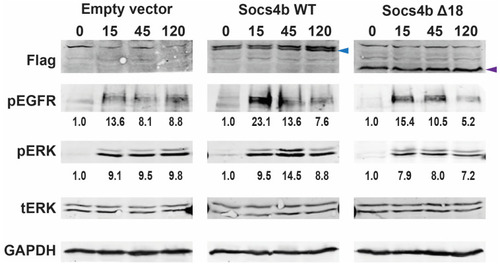
Analysis of the impact of zebrafish Socs4b on EGFR signaling. HEK293T cells transfected with empty vector control or plasmids encoding Flag-tagged Socs4b WT or Socs4b Δ18 were stimulated with EGF for the time indicated (min). Protein was extracted and subjected to Western blot analysis with antibodies against the Flag-tag to confirm expression of the Socs4b forms, as well as to pEGFR and pERK to detect EGFR signaling, along with tERK and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) that served as loading controls. The position of Flag-tagged Socs4b WT and Socs4b Δ18 proteins are shown with blue and purple arrowheads, respectively. Bands were quantified and the relative ratio of pEGFR:GAPDH and pERK:tERK calculated and shown below the relevant panel, with the 0 min sample set at 1.0 in each case, showing the average of two independent replicates. Original blots can be found in |