- Title
-
A modular chemigenetic calcium indicator for multiplexed in vivo functional imaging
- Authors
- Farrants, H., Shuai, Y., Lemon, W.C., Monroy Hernandez, C., Zhang, D., Yang, S., Patel, R., Qiao, G., Frei, M.S., Plutkis, S.E., Grimm, J.B., Hanson, T.L., Tomaska, F., Turner, G.C., Stringer, C., Keller, P.J., Beyene, A.G., Chen, Y., Liang, Y., Lavis, L.D., Schreiter, E.R.
- Source
- Full text @ Nat. Methods
|
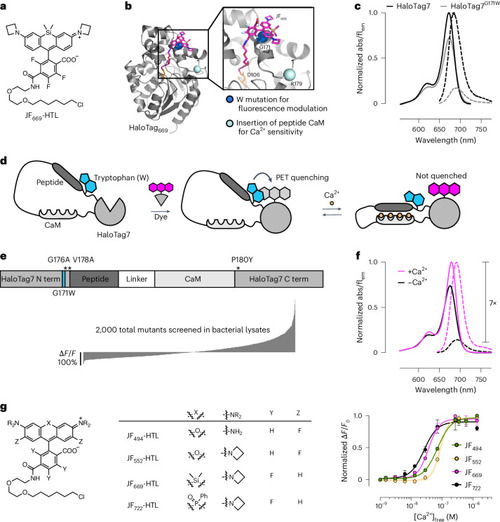
Engineering chemigenetic Ca2+ indicators with tryptophan quenching. |
|
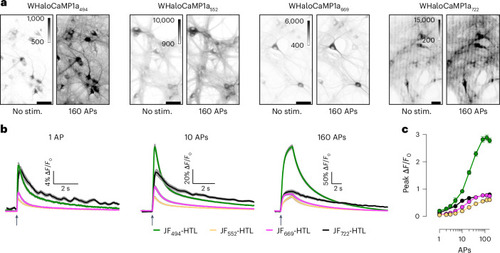
Characterization of WHaloCaMP1a in neuronal cultures. |
|
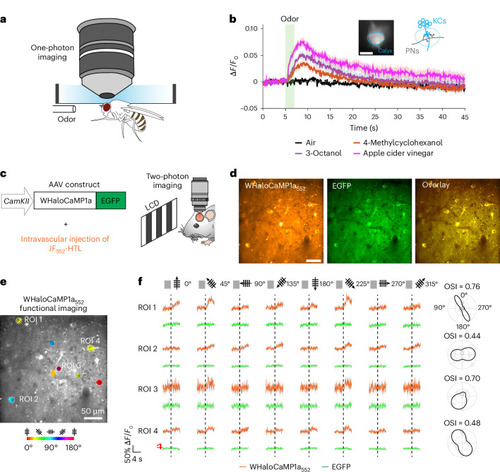
WHaloCaMP1a reports on neuronal activity in flies and mice. |
|
Three-color multiplexed functional imaging in zebrafish larvae. |
|
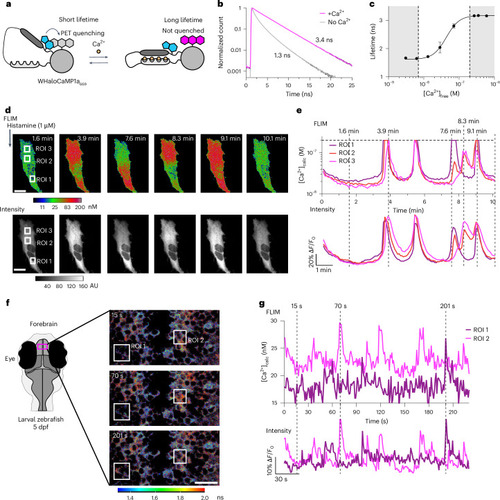
Quantitative Ca2+ measurements by FLIM using WHaloCaMP1a. |