|
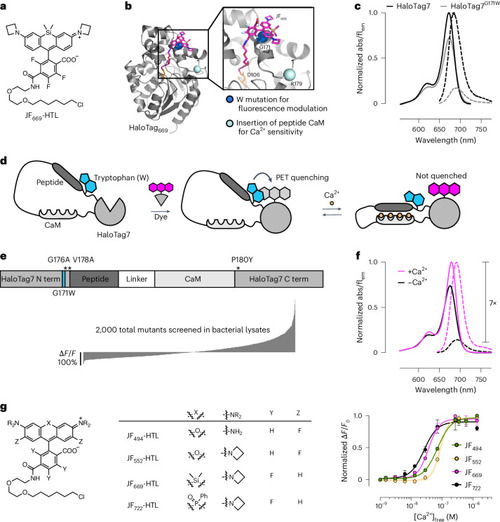
Engineering chemigenetic Ca2+ indicators with tryptophan quenching. a, Chemical structure of the JF669-HaloTag ligand (HTL). b, Crystal structure of HaloTag7 bound to the JF669-HaloTag ligand (HaloTag669) (PDB 8SW8). The positions of G171, which was mutated to a tryptophan to quench dye fluorescence emission, and R179, where Ca2+-sensitive protein domains were inserted, are highlighted as spheres. c, Normalized absorption (abs; solid lines) and fluorescence emission (flem; dashed lines) spectra of the JF669-HaloTag ligand bound to HaloTag7 or the HaloTag7G171W mutant. d, Schematic representation of WHaloCaMP, showing domain arrangement, covalent binding of the dye-ligand and the quenching tryptophan. e, Primary structure of WHaloCaMP1a (top) and ∆F/F0 values of variants (bottom) from a bacterial lysate screen to select WHaloCaMP1a. Term, terminus. f, Normalized absorption (solid lines) and fluorescence emission (dashed lines) spectra of the JF669-HaloTag ligand bound to purified WHaloCaMP1a in the presence (magenta) and absence (black) of Ca2+. g, Chemical structures of the dye-ligands used here with WHaloCaMP1a (left) and normalized Ca2+ titrations of WHaloCaMP1a bound to these dye-ligands (right). Data points represent mean and s.d. from n = 3 replicates. [Ca2+], Ca2+ concentration.
|