- Title
-
Creg1 Regulates Erythroid Development via TGF-β/Smad2-Klf1 Axis in Zebrafish
- Authors
- Han, X., He, W., Liang, D., Liu, X., Zhou, J., de Thé, H., Zhu, J., Yuan, H.
- Source
- Full text @ Adv Sci (Weinh)
|
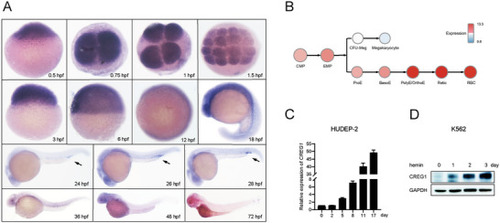
The expression profile of |
|
|
|
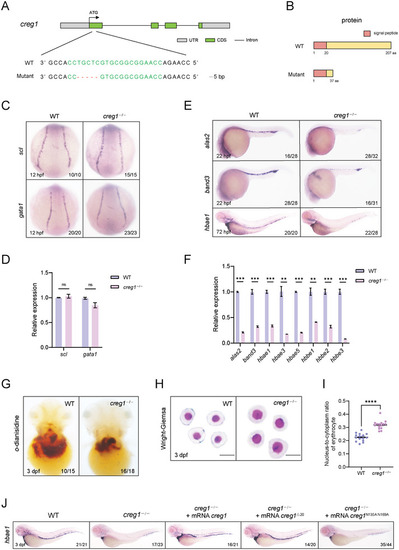
Loss of |
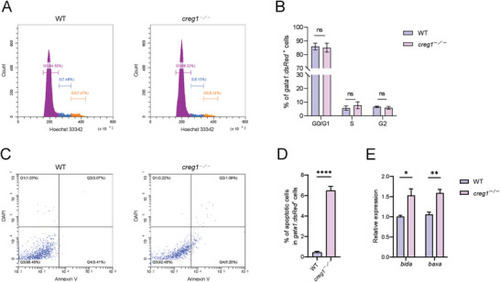
|
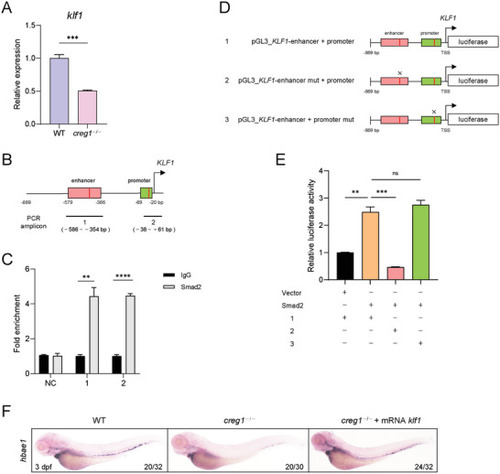
TGF‐β/Smad2 signaling pathway is implicated in |
|
|