- Title
-
pIGLET: Safe harbor landing sites for reproducible and efficient transgenesis in zebrafish
- Authors
- Lalonde, R.L., Wells, H.H., Kemmler, C.L., Nieuwenhuize, S., Lerma, R., Burger, A., Mosimann, C.
- Source
- Full text @ Sci Adv
|
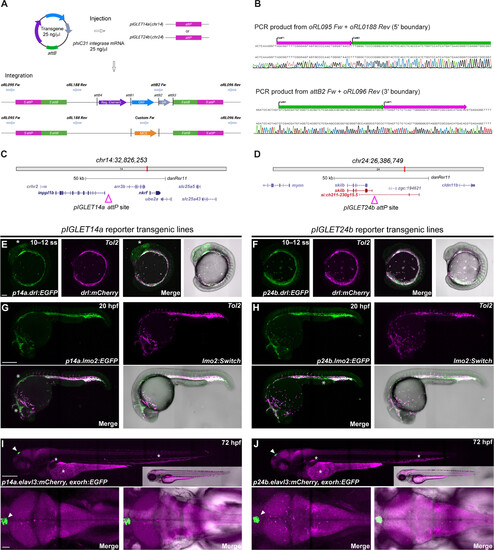
Zebrafish pIGLET14a and pIGLET24b landing sites enable high-quality transgenics. (A and B) Schematic of pIGLET system workflow. Transgenes with attB-containing pDEST or MCS backbones are coinjected with phiC31 integrase mRNA into pIGLET14a or pIGLET24b zebrafish (A). Integration is depicted with both attB-containing pDEST (pCM268, pRL055, pRL056, pCK122, and pCK123) (top) or MCS (pRL092 and pRL093) (bottom) backbones (A). The 5′ and 3′ transgene integration boundaries can be confirmed via PCR and sequencing (B). (C and D) Genomic locations of both landing sites. (E to J) Validating pIGLET14a and pIGLET24b with F2 drl:EGFP, lmo2:EGFP,cryaa:Venus and elavl3:mCherry,exorh:EGFP. (E and F) Crossed to Tol2-based drl:mCherry, p14a.drl:EGFP and p24b.drl:EGFP show analogous reporter activity in lateral plate mesoderm (LPM; 10 to 12 ss green fluorescence). Note faint brain expression in p14a.drl:EGFP (E, white asterisk) common with Tol2-based drl reporter transgenics with strong expression. (G and H) Crossed to Tol2-based lmo2:Switch (dsRed2, magenta), p14a.lmo2:EGFP and p24b.lmo2:EGFP show analogous, overlapping activity in endothelium at 20 hpf. EGFP expression is more consistent/complete compared to dsRED2 in Tol2-based lmo2:Switch (G and H, white asterisk). (I and J) p14a.elavl3:mCherry and p24b.elavl3:mCherry show complete, consistent reporter expression in the central nervous system at 72 hpf, akin to previous Tol2-based elavl3 transgenics. The in cis exorh:EGFP transgenesis marker indicated with white arrowhead. Note common autofluorescence in blood, kidney, and yolk (I and J, white asterisk). F2 heterozygous embryos are depicted (E to J). Scale bars: 100 μm (E), 250 μm (G), 250 μm (I), and 50 μm. |
|
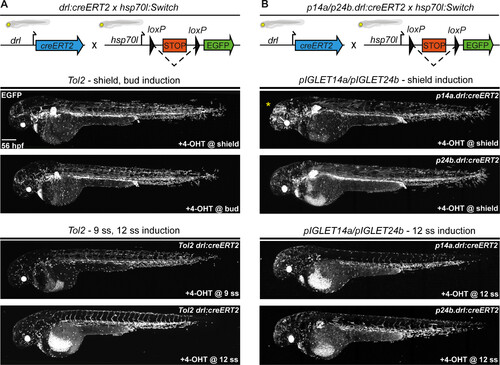
CreERT2 driver transgenes have predictable and consistent activity in pIGLET14a and pIGLET24b. Comparison of CreERT2-mediated recombination of Tol2-based drl:creERT2 and pIGLET-based lines p14a.drl:creERT2 and p24b.drl:creERT2 by crossing to the loxP reporter hsp70l:Switch and performing a 4-OHT induction series. Transgene and crossing schematics on top, lateral confocal views and anterior to the left. (A and B) Compared to the Tol2-based drl:creERT2 transgenic line, p14a.drl:creERT2 and p24b.drl:creERT2 show accurate lineage contribution dynamics at 56 hpf when treated with 4-OHT at shield stage and 12 ss (A and B). Note that with shield stage 4-OHT induction, drl-expressing cells contributed both to the LPM lineages (heart, pectoral fin, endothelial cells, kidney, mesothelia, blood, pharyngeal arch musculature, and macrophages) and a subset of paraxial mesoderm lineages (median fin fibroblasts and skeletal muscle). With 12 ss 4-OHT induction, LPM lineages are specifically traced with more mosaic labeling of heart, pectoral fin, and kidneys (A and B). Consistent with our p14a.drl:EGFP reporter lines, some neuronal lineage labeling is observed with p14a.drl:creERT2 when induced with 4-OHT at shield stage (yellow asterisk) (B). F2 heterozygous embryos are depicted (B). Scale bar, 200 μm. |
|
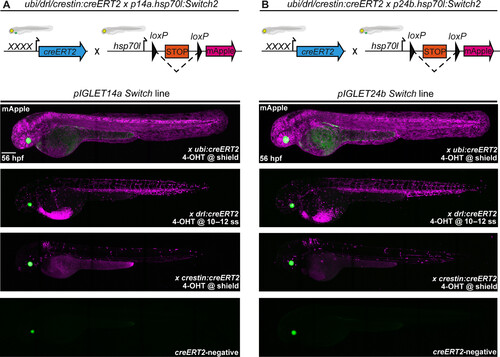
loxP Switch transgenes have predictable and consistent activity in pIGLET14a and pIGLET24b. Comparison of CreERT2-mediated recombination of pIGLET-based loxP reporter hsp70l:Switch2. Transgene and crossing schematics on top, lateral confocal views and anterior to the left. (A and B) Testing of p14a.hsp70l:Switch2 and p24b.hsp70l:Switch2 with three independent CreERT2-driver lines: ubi:creERT2, drl:creERT2, and crestin:creERT2. p14a.hsp70l:Switch2 and p24b.hsp70l:Switch2 drive strong mApple reporter expression following CreERT2-mediated recombination when crossed to ubiquitous and tissue-specific CreERT2-driver lines, treated with 4-OHT, and imaged at 56 hpf (A and B). Note the near-complete mApple labeling of the embryo at 56 hpf when p14a.hsp70l:Switch2 and p24b.hsp70l:Switch2 are combined with ubi:creERT2 and accurate labeling of the LPM or neural crest cells when used in combination with drl:creERT2 or crestin:creERT2, respectively, akin to previous results using Tol2-based versions of either CreERT2 driver (A and B). F2 heterozygous embryos are depicted (A and B). Scale bar, 200 μm. |
|
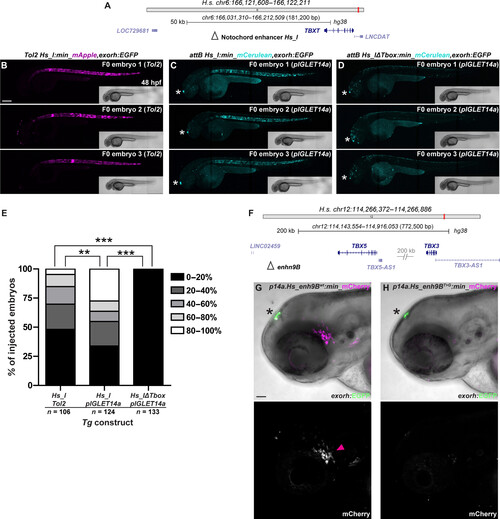
pIGLET-based transgenesis facilitates reproducible enhancer variant testing in Zebrafish. (A to E) F0 injection–based testing of notochord enhancer Hs_I using Tol2- and pIGLET-based transgenesis. Schematic of TBXT locus with Hs_I annotated (A). Confocal imaging for mApple (B) or mCerulean (C and D) with bright-field inserts, anterior to the left. Both Tol2- and pIGLET14a-based F0 injections of Hs_I:min-mApple/mCerulean,exorh;EGFP result in embryos showing reporter expression with 80 to 100% fluorescent notochord coverage (B and C). Removal of the Tbox motif from Hs_I results in a complete loss of notochord reporter activity (D). Injected zebrafish are sorted for the exorh:EGFP transgenesis marker (white asterisk) to confirm successful injection (C and D). pIGLET14a-based F0 injections of Hs_I:min-mCerulean,exorh:EGFP show significantly more embryos with 80 to 100% fluorescent notochord coverage compared to Tol2-based injections and the Tbox motif deletion variant (E). (F to H) Comparison of stable reporter lines driven by the human TBX5/TBX3-associated enhancer 9B (Hs_enh9B, wt versus T > G variant). Schematic of TBX5/3 locus with enh9B annotated (F). Hs_enh9Bwt drives reporter expression in the ventral lateral line ganglia (magenta arrowhead) (G). Compared to wt, the disease-associated variant Hs_enh9BT>G exhibits nearly a complete loss of mCherry reporter expression (G and H). F1 heterozygous embryos are depicted (G and H). Scale bars: 200 μm (B) and 50 μm (G). |