- Title
-
Modulating mycobacterial envelope integrity for antibiotic synergy with benzothiazoles
- Authors
- Habjan, E., Lepioshkin, A., Charitou, V., Egorova, A., Kazakova, E., Ho, V.Q., Bitter, W., Makarov, V., Speer, A.
- Source
- Full text @ Life Sci Alliance
|
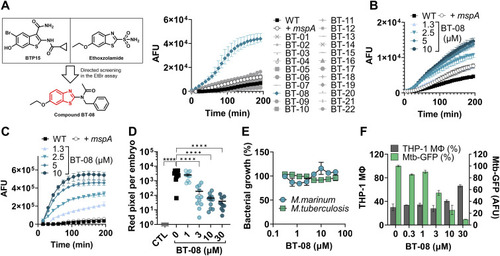
Screening for membrane permeabilizing agents identifies BT-08 as a hit compound with in vivo and ex vivo anti-mycobacterial activity. |
|
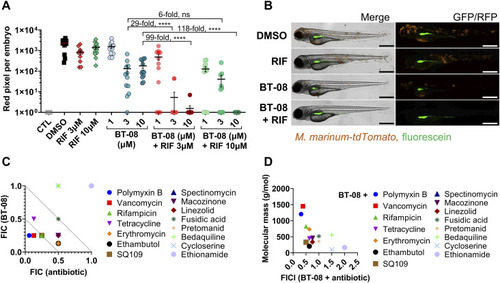
Compound BT-08 and high molecular weight antibiotics act synergistically in vivo and in vitro. |
|
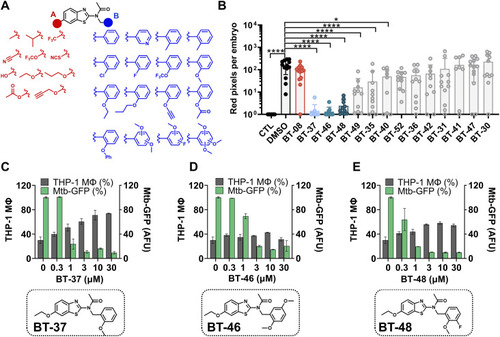
Structure–activity relationship studies revealed the most active derivative BT-37. |
|
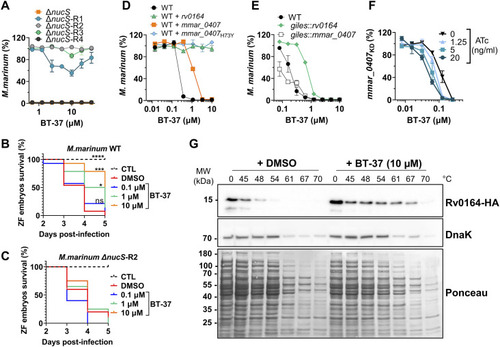
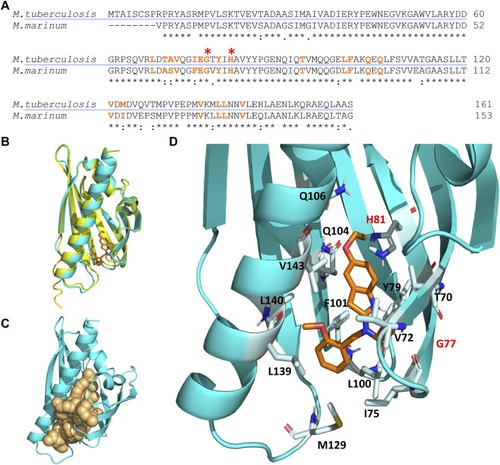
Mutations and expression level of MMAR_0407 (Rv0164) modulate susceptibility to BT-37. |
|
Molecular docking simulation of the structure of the Rv0164 complex with BT-37. |
|
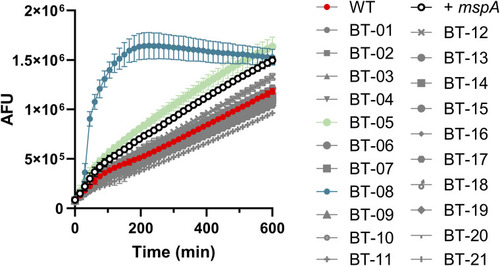
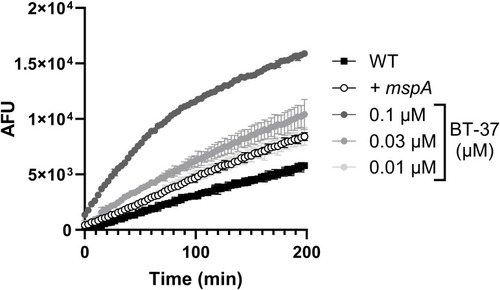
Compound BT-08 increases resazurin uptake. WT |
|
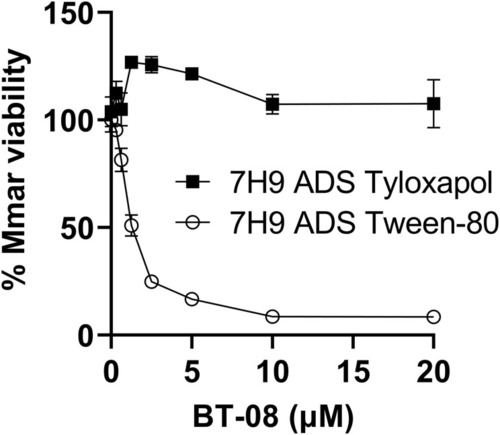
Detergents used in growth media affect the susceptibility of M. marinum to BT-08. Susceptibility of |
|
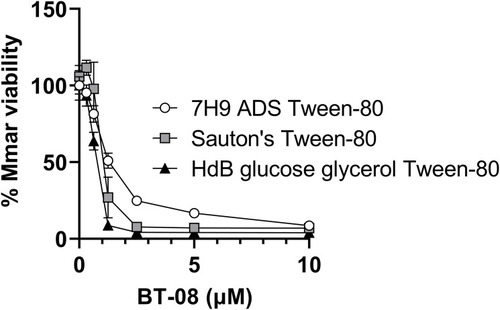
Different growth media affect susceptibility of M. marinum to BT-08. Susceptibility of |
|
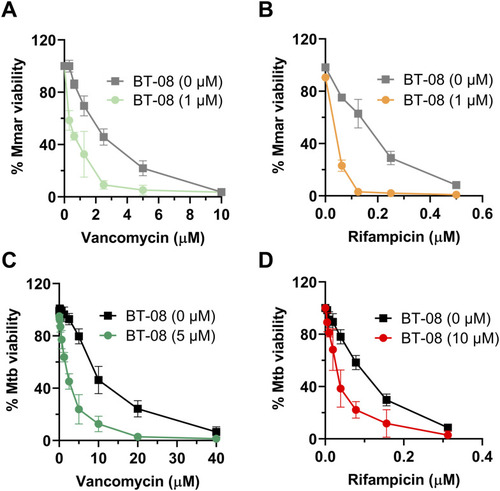
Compound BT-08 increases activity of rifampicin (RIF) and vancomycin in vitro. |
|
Treatment of M. marinum with BT-37 increases ethidium bromide (EtBr) uptake. The |
|
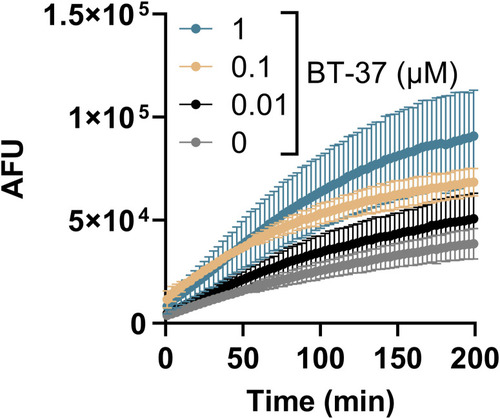
Treatment of M. tuberculosis with BT-37 increases ethidium bromide (EtBr) uptake. The |
|
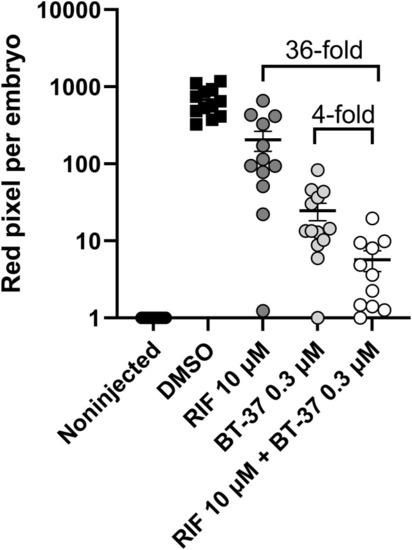
Synergistic effect of RIF and BT-37 in The activity of rifampicin at 10 μM and BT-37 at 0.3 μM and their combination in the zebrafish embryo infection model. Zebrafish embryos were yolk-infected with |
|
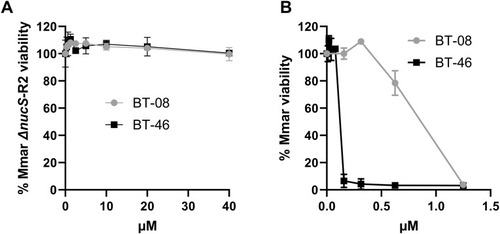
BT-08 and BT-46 exhibit cross-resistance to the BT-37–resistant M. marinum strain. |
|
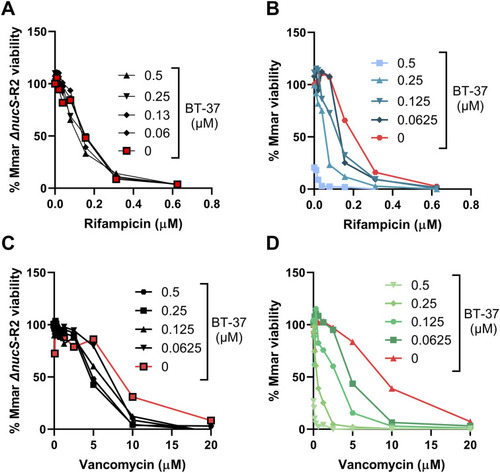
BT-37–resistant M. marinum strain does not show synergy with vancomycin or rifampicin in vitro |
|
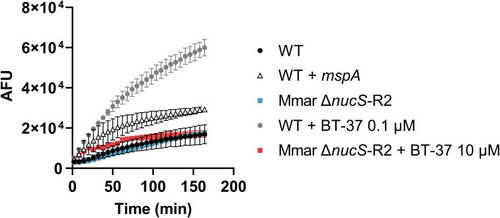
Treatment of BT-37–resistant M. marinum does not increases ethidium bromide (EtBr) uptake. The |
|
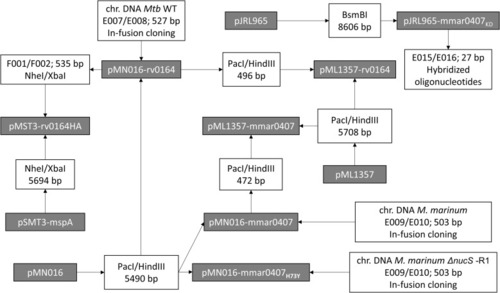
Cloning strategy for new plasmids used in this study. Constructed plasmids are included in gray boxes. The primer pairs used for PCR amplification and correlated restriction enzymes for cloning are listed in white boxes. The DNA template for the PCRs is listed above the primer pairs. If several primer pairs are listed, overlap PCR was used to fuse the PCR fragments. When a DNA fragment was obtained by digestion of a plasmid, the used restriction enzymes and the length of the obtained fragments are indicated. Constructed plasmids with their features and primers with their sequences are listed in Tables S9 and S10, respectively. |