- Title
-
Heritable CRISPR Mutagenesis of Essential Maternal Effect Genes as a Simple Tool for Sustained Population Suppression of Invasive Species in a Zebrafish Model
- Authors
- Krueger, C.J., Dai, Z., Zhu, C., Zhang, B.
- Source
- Full text @ Zebrafish
|
A heritable CRISPR mutagenesis strategy leading to sustained sex-specific disruption of reproduction as a simple and efficient method for population suppression in fish. Schematic of the HMEL strategy. Left: A CRISPR knockin cassette with ubiquitous expression of Cas9 and a gRNA targeting an essential maternal effect gene - here npm2b - is integrated in the genome. Right: Fish inheriting the knockin, called HMEL fish, express CRISPR nuclease causing npm2b mutant maternal embryo lethal phenotype (red) in females, while fertile HMEL males (yellow) can pass on the construct. HMEL, heritable maternal effect embryo lethality; npm2b, nucleoplasmin 2b; WT, wildtype. |
|
HMEL allele leads to robust mutation in the npm2b target and HMEL effect in female fish. (A) A high-activity CRISPR target site identified in npm2b exon 4. PAM underlined and AgeI restriction site in bold. (B) Somatic (caudal fin) mutagenesis of the npmb2 target site in individual mature F1 HMEL fish measured by AgeI disruption and quantified by densitometry. Average mutagenesis is 95.0% (±5.6% SD). (C) Fish carrying the HMEL allele develop as mosaic npm2b mutants. Gametes from an HMEL heterozygote may have one of four different genotypes, carrying either a WT or mutant npm2b allele each with or without the HMEL allele. After breeding with a WT fish, four different classes of offspring may be observed. Embryos not inheriting HMEL have a static genotype (either HMEL?/? npm2b+/+, or HMEL?/? npm2b+/?) throughout development. In contrast, HMEL embryos (HMEL+/?) experience dynamic genetic changes resulting in a varied mosaic cellular genotype of npm2b throughout development. (D) Reproduction of female and male F1 HMEL fish following outcross, measured as live embryos at 24 hpf as a fraction of total embryos counted at 2 hpf. Female HMEL embryo survival (0.21 ± 0.58%) was significantly less than WT cross by unpaired two-tailed t-test (****p < 10?7), while male HMEL embryo survival showed no significant difference compared with WT pairings (n.s.; p = 0.69). Data are shown as mean ± SD. (E) Upper panel: Representative image of offspring of female HMEL outcross at 24 hpf shows majority of embryos died, while surviving embryos showed notable developmental delay. Lower panel: Representative image of offspring of male HMEL outcross at 24 hpf shows normal development. Scale bar 1 mm. (F) Individual F2 embryos inheriting the HMEL allele (HMEL+/?) from outcross of an HMEL male showed significant mutagenesis in the npm2b target at 6 dpf compared with noninheriting sibling embryos (HMEL?/?) (ANOVA, p < 10?4), measured by AgeI restriction site disruption. ANOVA, analysis of variance; dpf, days postfertilization; hpf, hours postfertilization; SD, standard deviation. |
|
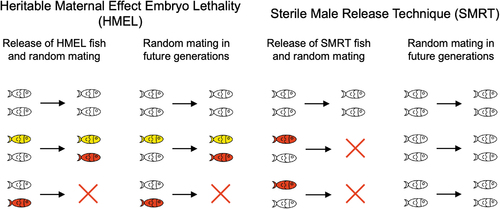
HMEL may lead to sustained population suppression, in contrast to SMRT. Comparison of HMEL and SMRT strategies. Left: With HMEL, the population suppression due to targeted females (red) may persist after the period of introduction through the fertile HMEL male (yellow) lineage. Later introductions can increase the HMEL allele frequency and further increase negative pressure on the population. Right: With SMRT, the sterility phenotype (red) is static and population suppression is limited to the life span of introduced modified individuals, and the target population may rapidly recover if new sterile individuals are not regularly introduced. SMRT, sterile male release technique |