- Title
-
Digenic inheritance involving a muscle-specific protein kinase and the giant titin protein causes a skeletal muscle myopathy
- Authors
- Töpf, A., Cox, D., Zaharieva, I.T., Di Leo, V., Sarparanta, J., Jonson, P.H., Sealy, I.M., Smolnikov, A., White, R.J., Vihola, A., Savarese, M., Merteroglu, M., Wali, N., Laricchia, K.M., Venturini, C., Vroling, B., Stenton, S.L., Cummings, B.B., Harris, E., Marini-Bettolo, C., Diaz-Manera, J., Henderson, M., Barresi, R., Duff, J., England, E.M., Patrick, J., Al-Husayni, S., Biancalana, V., Beggs, A.H., Bodi, I., Bommireddipalli, S., Bönnemann, C.G., Cairns, A., Chiew, M.T., Claeys, K.G., Cooper, S.T., Davis, M.R., Donkervoort, S., Erasmus, C.E., Fassad, M.R., Genetti, C.A., Grosmann, C., Jungbluth, H., Kamsteeg, E.J., Lornage, X., Löscher, W.N., Malfatti, E., Manzur, A., Martí, P., Mongini, T.E., Muelas, N., Nishikawa, A., O'Donnell-Luria, A., Ogonuki, N., O'Grady, G.L., O'Heir, E., Paquay, S., Phadke, R., Pletcher, B.A., Romero, N.B., Schouten, M., Shah, S., Smuts, I., Sznajer, Y., Tasca, G., Taylor, R.W., Tuite, A., Van den Bergh, P., VanNoy, G., Voermans, N.C., Wanschitz, J.V., Wraige, E., Yoshimura, K., Oates, E.C., Nakagawa, O., Nishino, I., Laporte, J., Vilchez, J.J., MacArthur, D.G., Sarkozy, A., Cordell, H.J., Udd, B., Busch-Nentwich, E.M., Muntoni, F., Straub, V.
- Source
- Full text @ Nat. Genet.
|
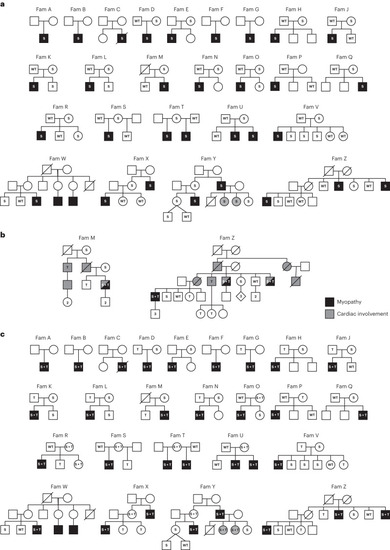
Pedigrees of the |
|
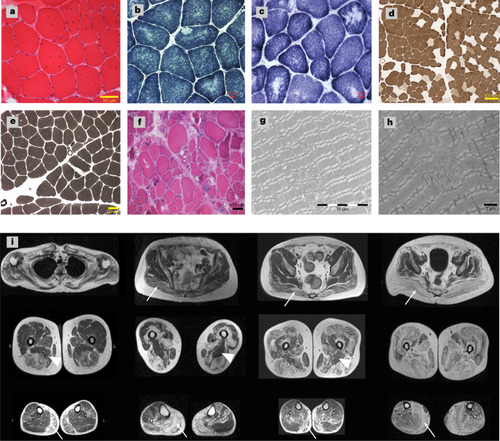
Muscle pathology of the patients with |
|
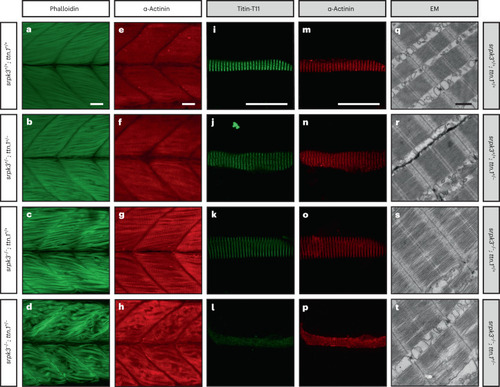
Titin immunoanalysis of patients with Muscle biopsy lysates of individuals DI:1, DII:1, LII:1, XII:3, XIII:1 and YII:3 were analyzed using different anti-titin antibodies. |
|
|
|
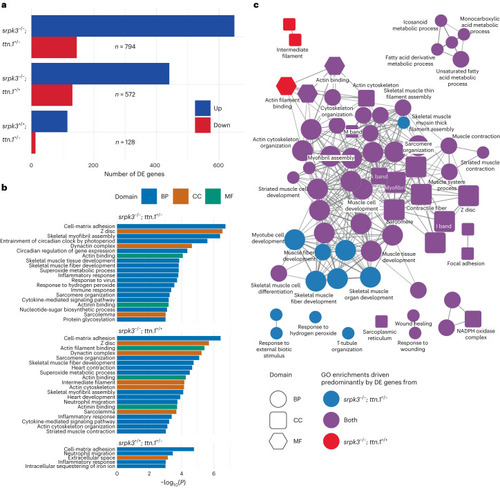
Transcriptome analysis of mutant zebrafish larvae. |
|
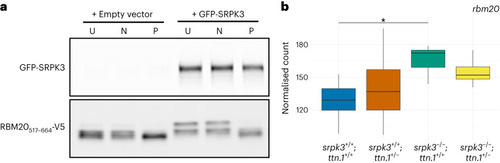
SRPK3 phosphorylates RBM20 in vitro. |