- Title
-
Conservation of affinity rather than sequence underlies a dynamic evolution of the motif-mediated p53/MDM2 interaction in ray-finned fishes
- Authors
- Mihalič, F., Arcila, D., Pettersson, M.E., Farkhondehkish, P., Andersson, E., Andersson, L., Betancur-R, R., Jemth, P.
- Source
- Full text @ Mol Bio Evol
|
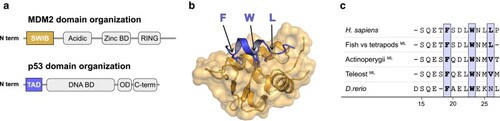
Structure of the human MDM2/p53TAD complex. a) Schematic domain architecture of MDM2 and p53. b) Crystal structure of the human complex between the SWIB domain of MDM2 (gold) and a peptide corresponding to the conserved binding motif in p53TAD (blue) (PDBid: 1ycr) ( |
|
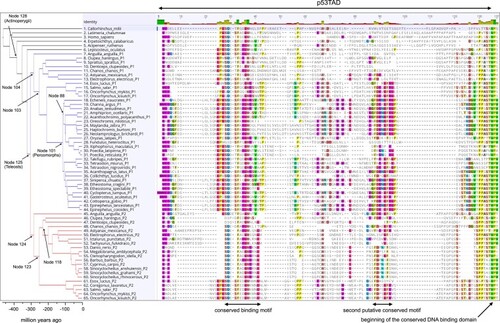
Sequence alignment of p53TAD mapped onto the fish phylogeny. We included 51 extant species representing some of the major lineages of fishes and for which high-quality sequencing data were available. The blue and red branches represent the split between the 2 paralogs p53TADP1 and p53TADP2, respectively, in teleosts (node 125). After the split, p53TADP1 and p53TADP2 have evolved in parallel in each lineage. Node numbers refer to the ancestral reconstruction analysis ( |
|
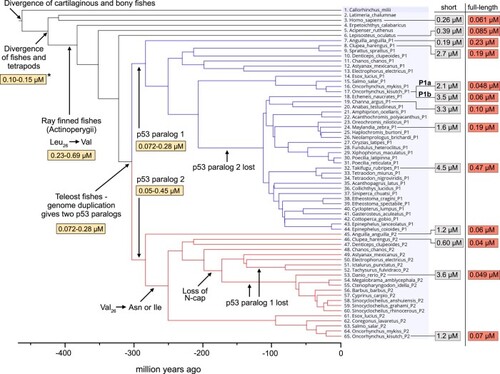
Affinities of p53TAD/MDM2 interactions. Affinities ( |
|
Affinities between p53TAD peptides and MDM2 from |
|
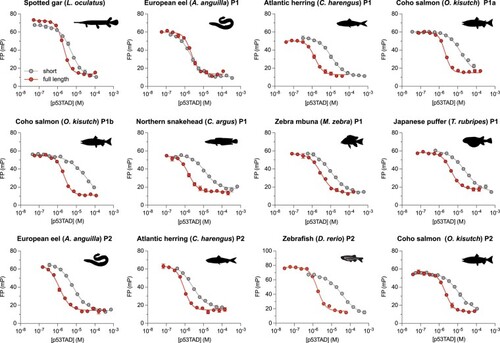
Affinity of short and full-length p53TAD of different extant fishes measured by fluorescence polarization competition experiments. Affinity is higher for full-length p53TAD constructs ( |