- Title
-
Molecular mechanism governing RNA-binding property of mammalian TRIM71 protein
- Authors
- Shi, F., Zhang, K., Cheng, Q., Che, S., Zhi, S., Yu, Z., Liu, F., Duan, F., Wang, Y., Yang, N.
- Source
- Full text @ Science Bull.
|
Structure of mTRIM71-NHL complexed with Trincr1 RNA fragment. (a) EMSA results of mTRIM71-NHL bound to different RNA motifs of Trincr1 containing predicted hairpin structure (F0 and F1) or triple-adenosine bases (F2 and F3). Trincr1-F0 was shifted to a constant position by mTRIM71-NHL at low protein to RNA molar ratio, but Trincr1-F1, F2 and F3 was hardly shifted by mTRIM71-NHL. (b) Structure of mTRIM71-NHL bound to a 11 nt Trincr1 RNA (colored by green and orange), and its superposition to the structure of DrLin41-NHL-Lin29A (colored by gray). A top view of the electrostatic potential surface of mTRIM71-NHL and Trincr1 RNA was shown on the right panel. (c) Enlarged view of detailed interactions between mTRIM71-NHL and Trincr1 RNA. Sidechains of interacting residues and nucleotides are highlighted in a stick model, and hydrogen bonds are presented as dotted lines. (d) Detailed interactions of mTRIM71-NHL with AI, AII and AIII of the RNA individually. (e) Schematic diagram of interactions between mTRIM71-NHL and Trincr1 RNA. Hydrogen bonds are presented as dotted lines, and π-π stacking interaction is shown as solid line. |
|
Key residues of mTRIM71-NHL for RNA-binding. (a) EMSA results of 11 nt Trincr1 RNA bound to wild-type mTRIM71-NHL or mutants (WT, R612A, R642A, K659A, D660A, Y689D, Y689A, R707A, R738A, R783A and R801A). Trincr1 RNA was similarly shifted to a constant position by wild-type and D660A and R707A mutants. Other mutants showed varying degrees of defects in RNA-binding properties. (b) FP results quantified the binding affinities of wild-type mTRIM71-NHL or mutants to the 11 nt Trincr1 RNA. KD values were shown and error bars were SEM. Six technical replicates (n = 6) were measured for the binding reaction of the wild-type protein, and three replicates (n = 3) were measured for each of the mutants. |
|
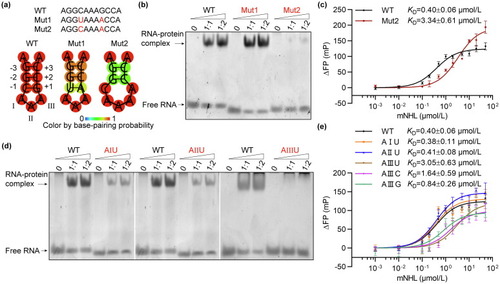
RNA preference of mTRIM71 protein. (a) Sequences and predicted secondary structures of wild-type 11 nt Trincr1 RNA and its structural mutants (Mut1 and Mut2). (b) EMSA results of mTRIM71-NHL bound to 11 nt Trincr1 RNA or its mutants. Mut1 RNA was shifted to a constant position by mTRIM71-NHL similar to the wild-type RNA, while Mut2 RNA was hardly shifted by mTRIM71-NHL at high protein to RNA molar ratio. (c) FP results quantified the binding affinity of Mut2 RNA to mTRIM71-NHL. KD values were shown and error bars were SEM. Three technical replicates (n = 3) were measured for the binding reaction of Mut2. (d) EMSA results of mTRIM71-NHL bound to 11 nt Trincr1 RNA or its sequential mutants (AIU, AIIU, and AIIIU). AIIIU exhibited an apparent defect in mTRIM71-NHL recruiting. (e) FP results quantified the binding affinities of the sequential mutants of RNA to mTRIM71-NHL. KD values were shown and error bars were SEM. Three technical replicates (n = 3) were measured for each binding reaction of the mutants. |
|
Mouse TRIM71 recognizes mRNA UTRs of other cell-cycle related genes. (a) Sequences and predicted secondary structures of RNA fragments deduced from UTRs of Cdkn1a, Rbl2 and Dbn1. (b) EMSA results of mTRIM71-NHL bound to RNA fragments of Trincr1, Cdkn1a-3′UTR, Cdkn1a-5′UTR, Rbl2-3′UTR and Dbn1-3′UTR. The predicted stem-loop RNA of Cdkn1a-3′UTR exhibits strong in vitro interactions with mTRIM71-NHL. (c) EMSA results of RNA fragments of Cdkn1a-3′UTR and Rbl2-3′UTR bound to mTRIM71-NHL and mutants. R642A, K659A and Y689A mutants severely weakened the RNA-protein recognition. (d) RIP-qPCR results of mESCs expressing wild-type, R642A, K659A and Y689A mutants of mTRIM71 with FLAG. Data were first normalized to the corresponding input and then to ESCs transfected with 3 × FLAG-GFP control. Enrichment of Cdkn1a and Rbl2 were observed, whereas Dbn1 was not evidently enriched. Malat1 was introduced as negative control. Values are means ± standard deviation (SD). n = 3 independent experiments. (****) P < 0.0001. |