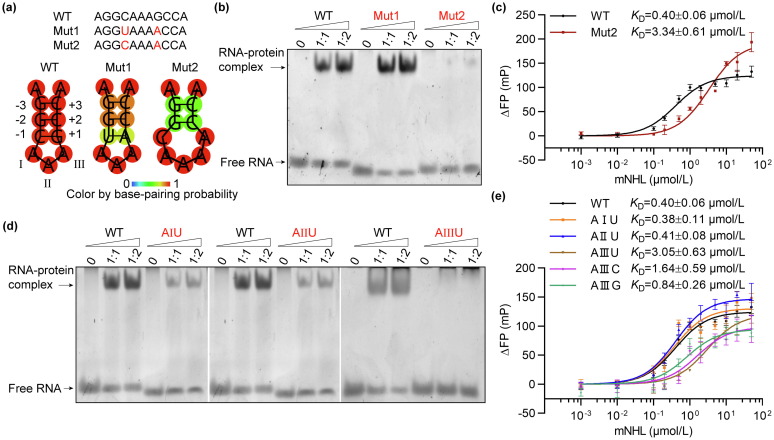
Fig. 3 RNA preference of mTRIM71 protein. (a) Sequences and predicted secondary structures of wild-type 11 nt Trincr1 RNA and its structural mutants (Mut1 and Mut2). (b) EMSA results of mTRIM71-NHL bound to 11 nt Trincr1 RNA or its mutants. Mut1 RNA was shifted to a constant position by mTRIM71-NHL similar to the wild-type RNA, while Mut2 RNA was hardly shifted by mTRIM71-NHL at high protein to RNA molar ratio. (c) FP results quantified the binding affinity of Mut2 RNA to mTRIM71-NHL. KD values were shown and error bars were SEM. Three technical replicates (n = 3) were measured for the binding reaction of Mut2. (d) EMSA results of mTRIM71-NHL bound to 11 nt Trincr1 RNA or its sequential mutants (AIU, AIIU, and AIIIU). AIIIU exhibited an apparent defect in mTRIM71-NHL recruiting. (e) FP results quantified the binding affinities of the sequential mutants of RNA to mTRIM71-NHL. KD values were shown and error bars were SEM. Three technical replicates (n = 3) were measured for each binding reaction of the mutants.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Science Bull.