Fig. 3
- ID
- ZDB-FIG-240319-88
- Publication
- Shi et al., 2023 - Molecular mechanism governing RNA-binding property of mammalian TRIM71 protein
- Other Figures
- All Figure Page
- Back to All Figure Page
|
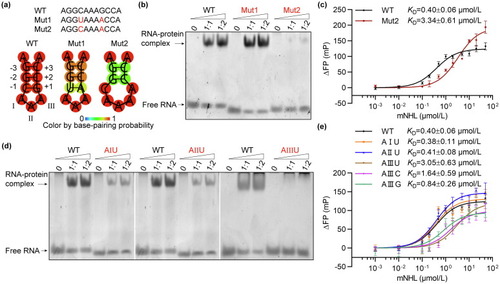
RNA preference of mTRIM71 protein. (a) Sequences and predicted secondary structures of wild-type 11 nt Trincr1 RNA and its structural mutants (Mut1 and Mut2). (b) EMSA results of mTRIM71-NHL bound to 11 nt Trincr1 RNA or its mutants. Mut1 RNA was shifted to a constant position by mTRIM71-NHL similar to the wild-type RNA, while Mut2 RNA was hardly shifted by mTRIM71-NHL at high protein to RNA molar ratio. (c) FP results quantified the binding affinity of Mut2 RNA to mTRIM71-NHL. KD values were shown and error bars were SEM. Three technical replicates (n = 3) were measured for the binding reaction of Mut2. (d) EMSA results of mTRIM71-NHL bound to 11 nt Trincr1 RNA or its sequential mutants (AIU, AIIU, and AIIIU). AIIIU exhibited an apparent defect in mTRIM71-NHL recruiting. (e) FP results quantified the binding affinities of the sequential mutants of RNA to mTRIM71-NHL. KD values were shown and error bars were SEM. Three technical replicates (n = 3) were measured for each binding reaction of the mutants. |