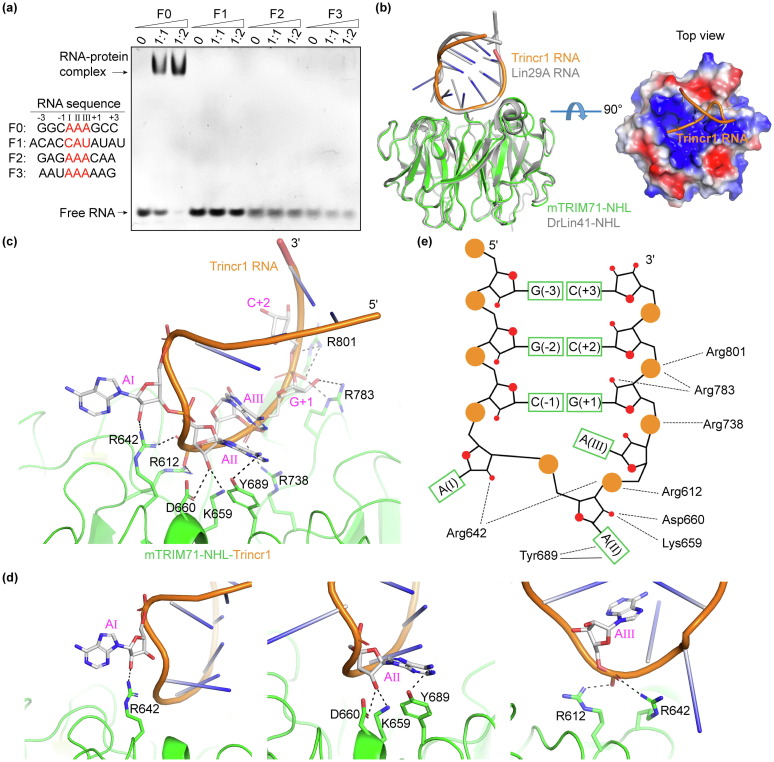
Fig. 1 Structure of mTRIM71-NHL complexed with Trincr1 RNA fragment. (a) EMSA results of mTRIM71-NHL bound to different RNA motifs of Trincr1 containing predicted hairpin structure (F0 and F1) or triple-adenosine bases (F2 and F3). Trincr1-F0 was shifted to a constant position by mTRIM71-NHL at low protein to RNA molar ratio, but Trincr1-F1, F2 and F3 was hardly shifted by mTRIM71-NHL. (b) Structure of mTRIM71-NHL bound to a 11 nt Trincr1 RNA (colored by green and orange), and its superposition to the structure of DrLin41-NHL-Lin29A (colored by gray). A top view of the electrostatic potential surface of mTRIM71-NHL and Trincr1 RNA was shown on the right panel. (c) Enlarged view of detailed interactions between mTRIM71-NHL and Trincr1 RNA. Sidechains of interacting residues and nucleotides are highlighted in a stick model, and hydrogen bonds are presented as dotted lines. (d) Detailed interactions of mTRIM71-NHL with AI, AII and AIII of the RNA individually. (e) Schematic diagram of interactions between mTRIM71-NHL and Trincr1 RNA. Hydrogen bonds are presented as dotted lines, and π-π stacking interaction is shown as solid line.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Science Bull.