- Title
-
Identification of side effects of COVID-19 drug candidates on embryogenesis using an integrated zebrafish screening platform
- Authors
- Ernst, A., Piragyte, I., Mp, A.M., Le, N.D., Grandgirard, D., Leib, S.L., Oates, A., Mercader, N.
- Source
- Full text @ Sci. Rep.
|
Overview of drugs included in zebrafish cardiovascular and behavioral screening setup. ( |
|
Schematic representation of the experimental workflow. ( |
|
Summary of results obtained for the compounds screening. ( |
|
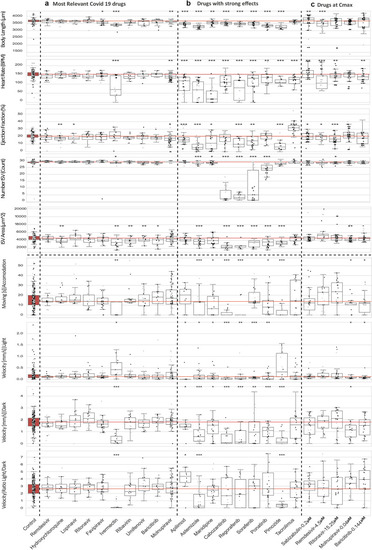
Combined boxplots and swarmplots showing the morphology and activity measurements for each larva. ( |
|
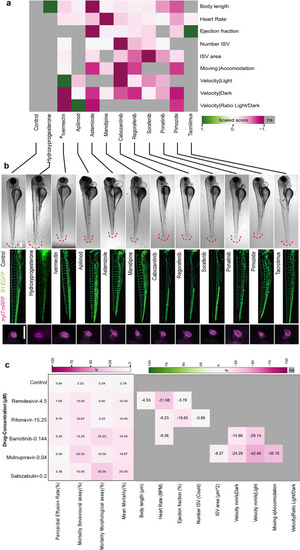
Selection of specific compounds eliciting effects on zebrafish embryonic development. ( |
|
qPCR results for inflammation associated gene expression in zebrafish larvae ( |
|
Remdesivir, Ritonavir, Molnupiravir, Baricitinib or Sabizabulin alter the inflammatory gene response in the absence of spike protein. qPCR results for inflammation associated gene expression in zebrafish larvae treaded with selected compounds. Remdesivir, Ritonavir, Molnupiravir, Baricitinib or Sabizabulin treatment is compared to larvae treated with DMSO. Bars represent mean and SD of 6 biological replicates. Each replicate consists of a pool of 5 larvae. Asterisks represent p-values from the Mann–Whitney U test * < 0.05; ** < 0.01;*** < 0.001. Note that in all graphs the same DMSO control samples were used. |
|
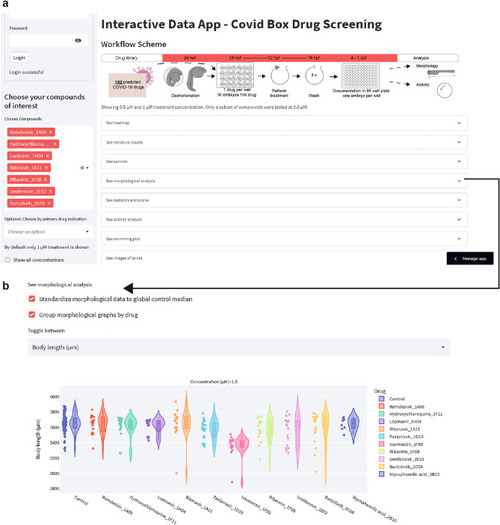
Generation of a Web-based Data App. ( |