- Title
-
Recruitment of transcription factor ETS1 to activated accessible regions promotes the transcriptional program of cilia genes
- Authors
- Zhang, D., Zhang, C., Zhu, Y., Xie, H., Yue, C., Li, M., Wei, W., Peng, Y., Yin, G., Guo, Y., Guan, Y.
- Source
- Full text @ Nucleic Acids Res.
|
|
|
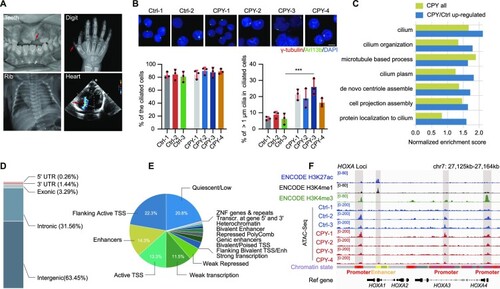
Open chromatin landscape remodelling occurs in EVC ciliopathy patients. ( |
|
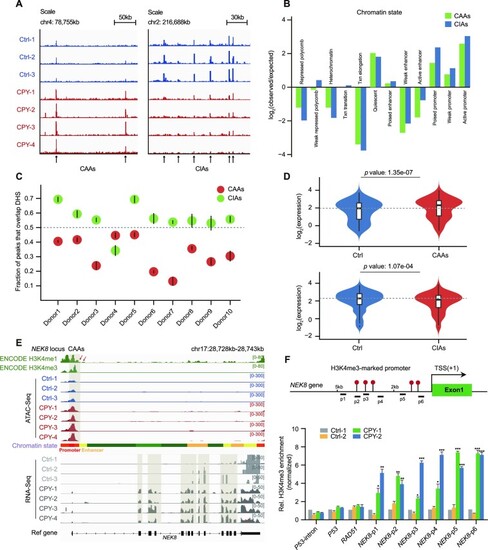
Genome-wide identification and characterization of EVC ciliopathy-related DARs. ( |
|
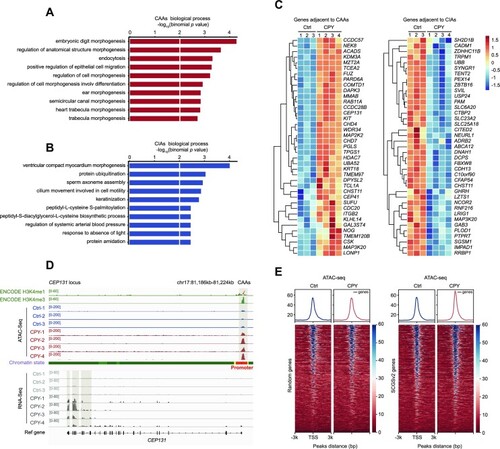
CAAs affect expression profiles of CAA-adjacent cilia genes. ( |
|
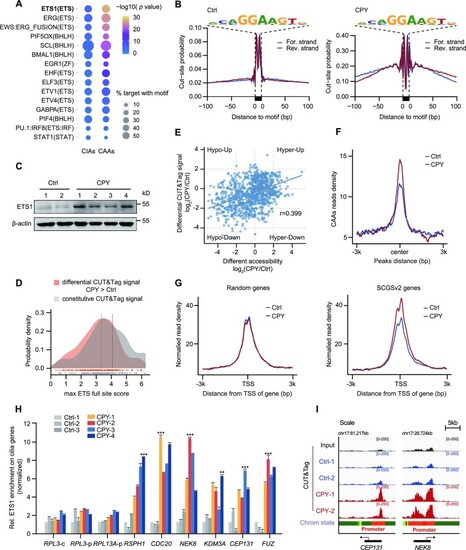
ETS1 is recruited to CAAs and leads to increased expression of cilia genes. ( |
|
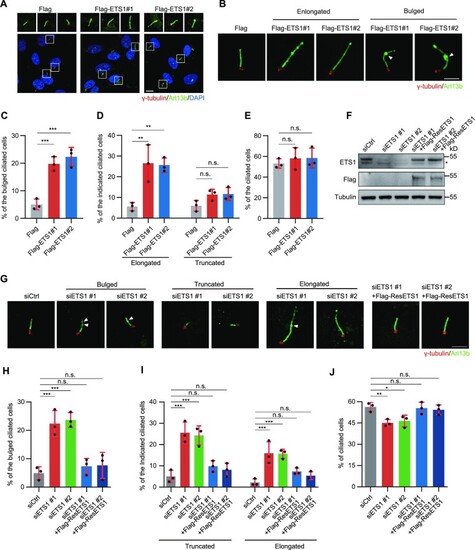
ETS1 positively regulates cilia formation. ( |
|
ETS1 loss induces aberrant expression of cilia genes by remodelling the signals of flanking CAAs. ( |
|
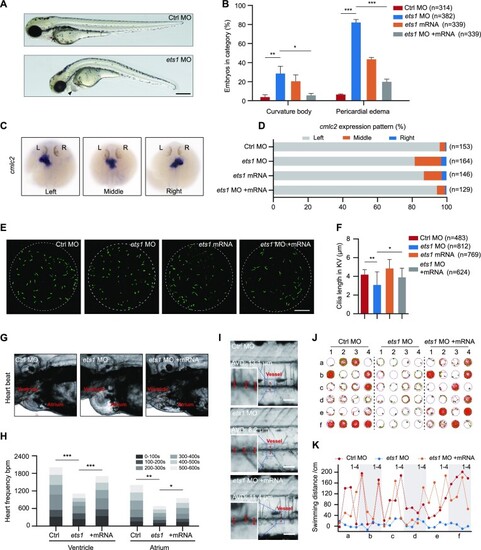
ETS1 expression alteration results in multiple ciliary defects in zebrafish larvae. ( |