- Title
-
The manipulation of cell suspensions from zebrafish intestinal mucosa contributes to understanding enteritis
- Authors
- Zhao, X., Liu, Y., Xie, J., Zhang, L., Zhu, Q., Su, L., Guo, C., Li, H., Wang, G., Zhang, W., Cheng, Y., Wu, N., Xia, X.Q.
- Source
- Full text @ Front Immunol
|
Operating processes for preparing cell suspension from zebrafish intestinal mucosa. |
|
Quality control of prepared intestinal single cell suspension from |
|
Enriching intestinal immune cells |
|
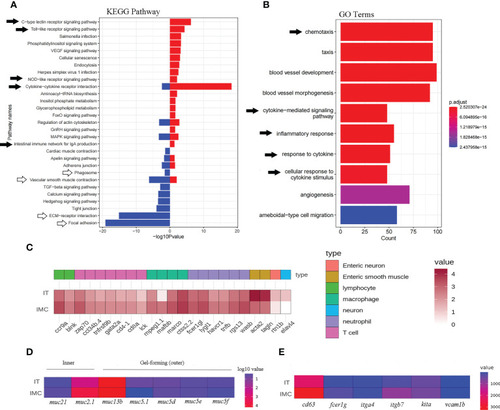
Enriched KEGG pathways and GO terms as well as heatmaps of mucosal immune related DEGs. Intestinal pathways |
|
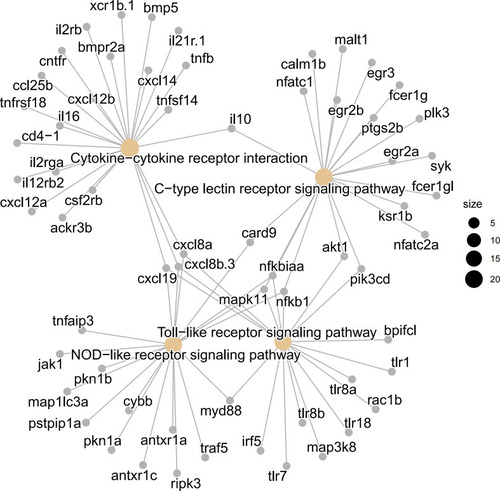
Visualization of involved genes in innate immune related KEGG pathways by cnetplot analysis. The pattern recognition related “C-type lectin receptor signaling pathway”, “NOD like receptor signaling pathway”, and “Toll like receptor signaling pathway”, as well as cytokine signaling related “cytokine-cytokine receptor interaction” were shown in details. |
|
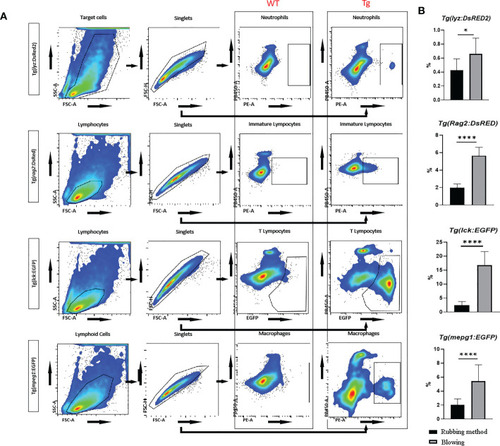
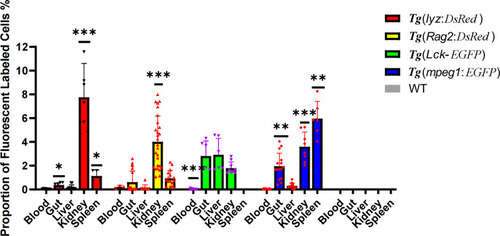
Flow cytometric analysis of transgenic zebrafish, including |
|
Proportion of fluorescent labeled immune cells, including neutrophils, macrophages, lymphocytes, and activated T cells, in immune organs (the periphery blood, intestine, liver, kidney and spleen). * represented |
|
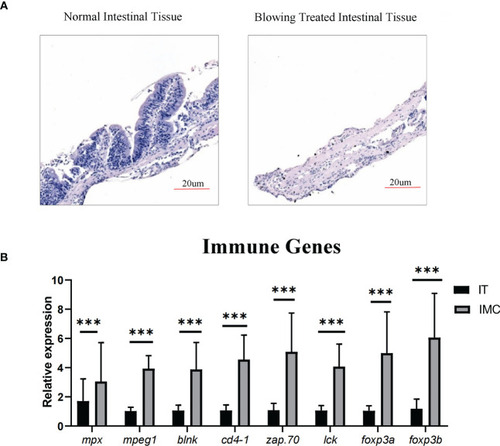
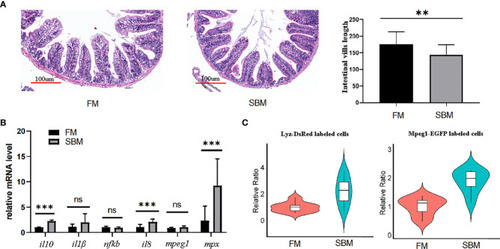
The SBMIE induced mucosa pathology was related to the altered composition of immune cells. The single cell suspension, obtained using current optimized method, accurately reflects altered composition of intestinal immune cells upon soybean induced inflammation. |