- Title
-
Gut-specific telomerase expression counteracts systemic aging in telomerase-deficient zebrafish
- Authors
- El Maï, M., Bird, M., Allouche, A., Targen, S., Şerifoğlu, N., Lopes-Bastos, B., Guigonis, J.M., Kang, D., Pourcher, T., Yue, J.X., Ferreira, M.G.
- Source
- Full text @ Nat Aging
|
Gut-specific and Cre-mediated tert expression rescues gut aging phenotypes.
a, Schematic representation of the transgene for Cre-inducible and tissue-specific expression of tert mRNA. b, RT–qPCR analysis of tert transgene mRNA and total tert mRNA (endogenous + transgene) expression in 9-month-old gut extracts (nWT = 5 and 6; Source data |
|
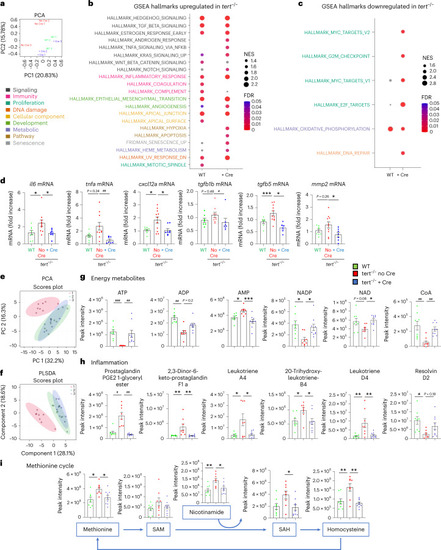
Gut-specific tert expression rescues gut transcriptomics and metabolomics profiles.
a, Principal component analysis (PCA)-based on untargeted transcriptomics data of 9-month-old gut samples. A clustering between tert−/− + Cre and WT was observed while the tert−/− no Cre group was clearly distinguishable from tert−/− no Cre fish (n = 3 per group). b,c, Identification of upregulated (b) or downregulated (c) hallmarks in tert−/− no Cre compared to either WT or tert−/− + Cre, based on GSEA. The normalized enrichment scores (NES) depict to what degree the pathway genes are overrepresented in WT or tert−/− + Cre, compared to tert−/− no Cre. Gene sets related to senescence, inflammation and morphogenesis were enriched while the hallmarks of proliferation and oxidative phosphorylation were downregulated in the gut of tert−/− no Cre fish compared to the other two groups. d, RT–qPCR analysis of inflammation-related gene expression (il6, tnfa, tgfb1b and tgfb5) and SASP-related gene expression (il6, tnfa, cxcl12a, tgfb1b, tgfb5 and mmp2) in 9-month-old gut samples (nWT = 8 fish, Source data |
|
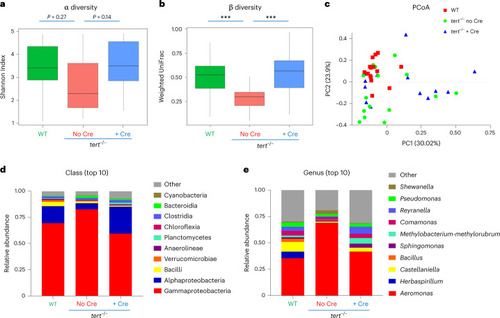
Gut-specific tert expression rescues gut microbiota dysbiosis.
Telomere elongation in the gut of tert−/− + Cre fish rescued gut microbiota composition and diversity to WT levels compared to tert−/− no Cre fish, which exhibited gut microbiota dysbiosis. a, Quantification of microbiome α diversity (within samples) using the Shannon index (P values were determined using a two-sided Wilcoxon signed-rank test) in the gut of 9-month-old fish. b, Quantification of microbiome β diversity using weighted UniFrac distance (within groups; ***P < 0.001 using a two-sided Tukey test) in the gut of 9-month-old fish. c, PCoA of the β diversity distance (weighted UniFrac) in the gut of 9-month-old fish. d, Relative abundance of top 10 bacterial classes in the microbiome of the three different groups in the gut of 9-month-old fish. e, Relative abundance of top 10 bacterial genera in the microbiome of the three different groups in the gut of 9-month-old fish. For all the figures, nWT = 15 fish, Source data |
|
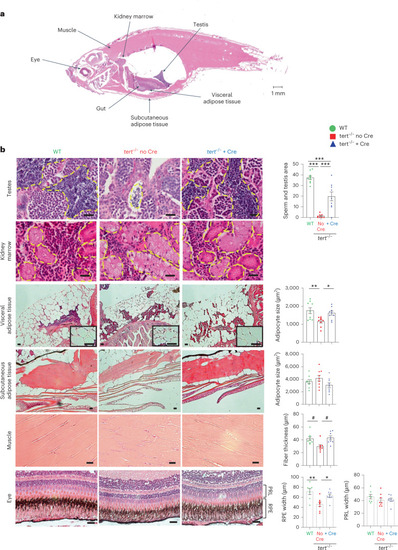
Gut-specific tert expression rescues systemic tissue degeneration.
Expression of telomerase in the gut of tert mutant fish rescued tissue degeneration in the testes, visceral adipose tissue, muscle and eye. a, Representative image of a longitudinal section of a zebrafish stained with H&E. The locations of each tissue analyzed in the study are indicated by arrows. b, Representative images of testes, kidney, visceral adipose tissue, subcutaneous adipose tissue, muscle and eye from 9-month-old fish stained with H&E (right). Except for the kidney, histological quantifications were performed for each tissue (left), namely the mature spermatids area (nWT = 10 fish, Source data |
|
Gut-specific tert expression rescues the aging phenotypes of testes.
a–e, Delaying gut aging in tert−/− + Cre fish rescues DNA damage, proliferation and senescence in the testes compared to tert−/− no Cre fish. a, Representative immunofluorescence images of DNA damage staining (γH2AX, left) and quantification (right, nWT = 6, Source data |
|
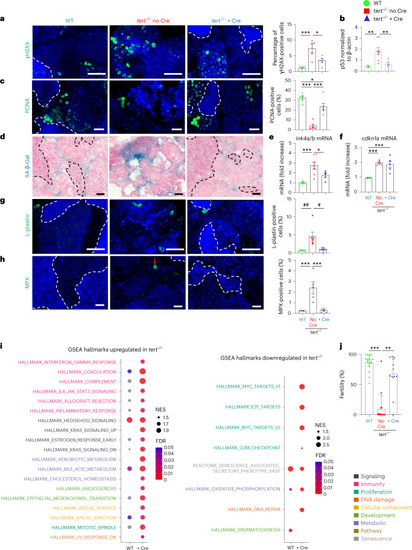
Gut-specific tert expression rescues aging of the hematopoietic system (kidney marrow).
a–f, Delaying gut aging in tert−/− + Cre fish rescues DNA damage, proliferation and senescence in the kidney marrow when compared to tert−/− no Cre fish. a, Representative immunofluorescence images of DNA damage staining (left, γH2AX) and quantification (right, nWT = 5, Source data |
|
Gut-specific tert expression extends the lifespan of tert−/− zebrafish.
Gut-specific telomerase activity extends the lifespan, increasing median life from 17 months in tert−/− no Cre fish to 24 months in tert−/− + Cre fish. The survival curve of WT (n = 42 fish), tert−/− no Cre (n = 38 fish) and tert−/− + Cre (n = 26 fish) zebrafish (**P < 0.01 using the log-rank test) is shown. Source data |
|
Gut-specific tert expression extends the healthspan of naturally aged zebrafish.
Expression of tert transgene in the gut of WT fish delays local aging phenotypes such as proliferation, senescence and tissue degeneration. This leads to beneficial systemic impact improving early aging phenotypes such as proliferation capacity. a, Representative immunofluorescence images of proliferation staining (left, PCNA) and quantification (right) in the gut, testes and kidney marrow of 27-month-old WT zebrafish expressing (WT + Cre; n = 7 fish for the gut and kidney marrow and n = 6 fish for the testes) or not expressing (WT no Cre; n = 8 fish for the gut and kidney marrow and n = 6 fish for the testes) tert transgene in the gut. b, Representative images of SA-β-Gal staining of gut, testes and kidney marrow sections of 24-month-old WT zebrafish expressing (WT + Cre) or not expressing (WT no Cre) the tert transgene in the gut (left). RT–qPCR analysis of senescence-associated genes ink4a/b (p15/16) and cdkn1a (p21) in the gut, testes and kidney marrow of either 9- (n = 6 fish) or 27-month-old WT zebrafish expressing (WT + Cre; n = 8 fish) or not expressing (WT no Cre; n = 8 fish) tert mRNA in the gut (right). c, Representative H&E-stained sections of the gut, testes and kidney marrow of 27-month-old WT zebrafish expressing (WT + Cre) or not expressing (WT no Cre) the tert transgene in the gut (left) and respective quantifications of the width of the gut lamina propria (right; n = 7 fish for WT no Cre and n = 6 fish for WT + Cre) and mature spermatid area (right; n = 7 fish for WT no Cre and WT + Cre). The yellow arrows indicate the width of the lamina propria quantified on the left. The dashed lines delineate the mature area of the spermatids. d, Survival curve of WT no Cre (n = 42 fish; similar to the WT curve in Fig. Fig.7)7) and WT + Cre (n = 36 fish) zebrafish. All data are presented as the mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, using a two-tailed unpaired t-test for a,c or a one-way ANOVA and post hoc Tukey tests for b; **P < 0.01, a using two-tailed Mann–Whitney U-test). All RT–qPCR graphs represent the mean ± s.e.m. mRNA fold increase after normalization by rps11 gene expression levels. Scale bar, 20 µm. Source data |