- Title
-
Calaxin stabilizes the docking of outer arm dyneins onto ciliary doublet microtubule in vertebrates
- Authors
- Yamaguchi, H., Morikawa, M., Kikkawa, M.
- Source
- Full text @ Elife
|
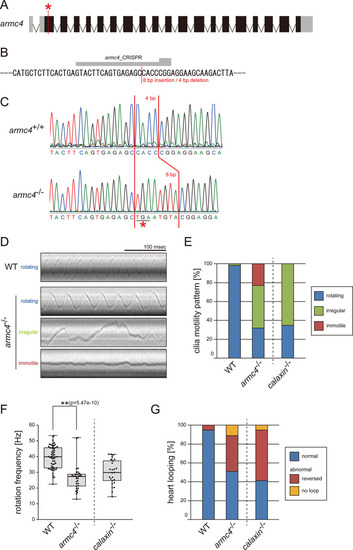
(A) Genomic organization of zebrafish armc4 gene. Black boxes: exons. Gray boxes: untranslated regions. Red asterisk: the genome-editing target site. (B) CRISPR/Cas9 target sequence. (C) Sanger sequencing of armc4+/+ and armc4−/− fish around the genome-editing target site. The 8 bp-insertion in armc4−/− includes a stop codon (red asterisk). (D) Typical kymographs of Kupffer’s vesicle cilia in WT and armc4−/− embryos. Kymograph patterns were categorized into three classes: rotating (blue), irregular (green), and immotile (red). Scale bar: 100 ms. (E) Ratios of each motility class. Number of cilia: 58 (WT) and 100 (armc4−/−). (F) Rotational frequencies of Kupffer’s vesicle cilia. Number of cilia: 58 (WT) and 32 (armc4−/−). Boxes correspond to the first and third quartiles, lines inside the boxes indicate the medians, and whiskers extend to the full range of the data. p-Value was calculated with Welch’s t-test. (G) Directions of heart looping. Number of embryos: 110 (WT) and 63 (armc4−/−). For comparison, calaxin-/- data from Sasaki et al., 2019 are displayed in (E, F, and G).
|
|
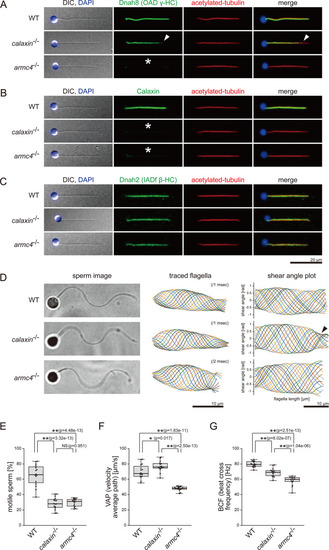
(A–C) Immunofluorescence microscopy of zebrafish spermatozoa. Scale bar: 20 μm. (A) Dnah8 was localized along the entire length of sperm flagella in WT. In calaxin-/-, Dnah8 was lost at the distal region of sperm flagella (white arrowheads). In arm4-/-, Dnah8 was lost (white asterisk). (B) Calaxin was localized along the entire length of sperm flagella in WT. In calaxin-/- and arm4-/-, Calaxin was lost (white asterisks). (C) Dnah2 was localized along the entire length of sperm flagella in WT, calaxin-/-, and arm4-/-. (D) Phase-contrast microscopy images of swimming spermatozoa (left column), traces of beating flagella (middle column), and shear angle plots of traced flagella (right column). Swimming spermatozoa were filmed using a high-speed camera at 1000 fps (frames per second). Shear angles were plotted against the distance from the flagellar base. In calaxin-/-, shear angle plots lost their slopes at the distal region of the flagella (black arrowhead). Scale bars: 10 μm. (E–G) Motilities of swimming spermatozoa were filmed using a high-speed camera at 200 fps and analyzed by CASA modified for zebrafish. For each zebrafish line, more than 600 spermatozoa were analyzed in total with 16 technical replicates. Spermatozoa with less than 20 μm/s velocities were considered immotile. (H) Ratio of motile spermatozoa. (I) Velocity of spermatozoa on averaged paths. The averaged paths were drawn by connecting the points of averaged sperm positions of contiguous 33 frames. (J) Frequencies at which sperm heads crossed their averaged paths. Boxes correspond to the first and third quartiles, lines inside the boxes indicate the medians, and whiskers extend to the full range of the data. p-Values were calculated with Tukey-Kramer test.
|
|
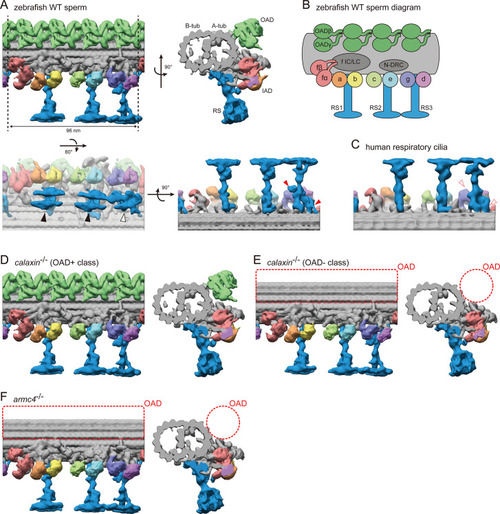
(A) DMT structure of WT zebrafish sperm flagella. A-tub and B-tub: A- and B-tubule of DMT, respectively. OAD: outer arm dynein, IAD: inner arm dynein, RS: radial spoke. Upper left: side view, upper right: base-to-tip view, lower left: bottom view, and lower right: back view of RSs. Black arrowheads indicate the spoke heads of RS1 and RS2. White arrowhead indicates the spoke head of RS3. Red arrowheads indicate the RS3 structures which are not found in the RS3 in human respiratory cilia. (B) Diagram of DMT structure. N-DRC: nexin-dynein regulatory complex. f IC/LC: IAD-f intermediate chain and light chain complex. (C) Back view of RSs in human respiratory cilia (EMD-5950; Lin et al., 2014). (D–E) DMT structures of calaxin-/- sperm flagella. Structural classification sorted the subtomograms into two classes: (D) OAD+ class (70.7%, 6122 particles) and (E) OAD- class (29.3%, 2535 particles). (F) DMT structure of armc4-/- sperm flagella. Green: OAD, pale red: IAD-f, orange: IAD-a, yellow: IAD-b, light-green: IAD-c, cyan: IAD-e, indigo: IAD-g, violet: IAD-d, and blue: RSs. Red circles indicate the loss of OAD.
|
|
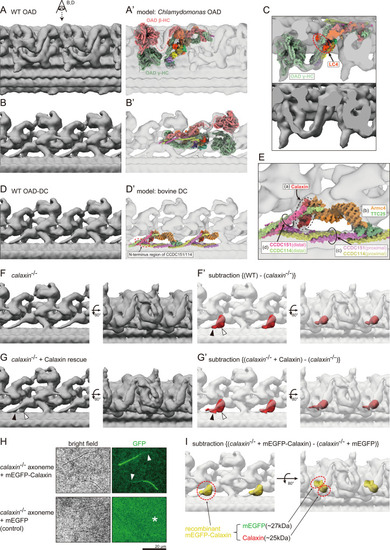
(A and B) OAD structure of WT zebrafish sperm flagella. Local refinement was performed focusing on OAD HCs. (B) shows the top view of A (eye and arrow). (A’ and B’) Comparison of zebrafish OAD structure with Chlamydomonas OAD model (PDB-7kzm; Walton et al., 2021). α-HC and DC linkers were omitted from the Chlamydomonas OAD model. (C) Detailed structure around OAD γ-HC. Upper: composite image of zebrafish OAD and Chlamydomonas OAD model. Lower: zebrafish OAD only. Chlamydomonas OAD has the LC4 protein (red circle) attached to the γ-HC tail. (D) OAD-DC structure of WT zebrafish sperm flagella. Local refinement was performed focusing on DC. (D’) Comparison of zebrafish DC structure with bovine DC model (PDB-7rro; Gui et al., 2021). Red dotted circle indicates the N-terminus region of the CCDC151/114. (E) Detailed structure of DC. DC is composed of four linker structures: (a) Calaxin, (b) the Armc4-TTC25 complex, (c) the proximal CCDC151/114, and (d) the distal CCDC151/114. (F) OAD-DC structure of calaxin-/- sperm flagella. (F’) Composite image of difference map (red; subtraction of F from WT) and calaxin-/- OAD-DC (translucent). Difference map shows the densities corresponding to the Calaxin (white arrowhead) and the adjacent CCDC151/114 (black arrowhead). (G) OAD-DC structure of calaxin-/- incubated with recombinant Calaxin protein. (G’) Composite image of difference map (red; subtraction of calaxin-/- from G) and calaxin-/- OAD-DC (translucent). (H) calaxin-/- sperm axoneme was incubated with recombinant proteins of mEGFP-Calaxin or mEGFP (control). Upper row: mEGFP-Calaxin binds to the limited region of calaxin-/- axoneme, with the partial loss of EGFP signals (white arrowheads). Lower row: mEGFP has no interaction with calaxin-/- axoneme (asterisk). Scale bar: 20 μm. (I) Composite image of difference map (yellow; subtraction of calaxin-/- incubated with mEGFP from calaxin-/- rescued by mEGFP-Calaxin) and calaxin-/- OAD-DC (translucent). Difference map shows the densities of mEGFP and Calaxin.
|
|
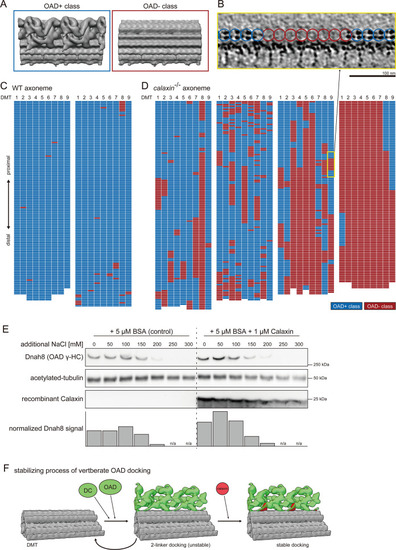
(A–D) Structural classification sorted the 24 nm repeat units of DMT into OAD+ class (blue) and OAD- class (red). (A) Averaged structures of each class. (B) Tomographic slice shows the side view of DMT. Blue or red circles indicate the class of each 24 nm repeat unit of DMT. Scale bar: 100 nm. (C–D) Distribution patterns of each class. 24 nm repeat units on each DMT are schematically displayed as individual colored grids. (C) Two typical tomograms of WT axoneme. (D) Four typical tomograms of calaxin-/- axoneme. Yellow square indicates the region displayed in B. (E) Immunoblot of calaxin-/- sperm axonemes incubated with or without recombinant Calaxin protein in different salt concentration buffers. Bottom row shows the bar graph of Dnah8 signals, normalized by the amount of acetylated tubulin signals. (F) Model of the stabilizing process of vertebrate OAD docking onto DMT.
|
|
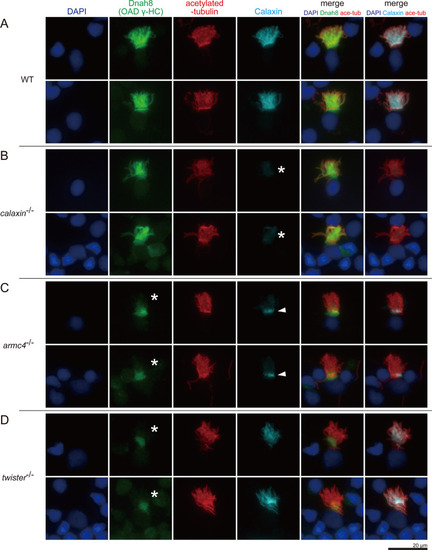
(A–D) Immunofluorescence microscopy of multiciliated cells of zebrafish olfactory epithelium. (A) WT. (B) calaxin-/-. Calaxin signal was lost (white asterisks). (C) armc4-/-. Ciliary localization of Dnah8 was lost (white asterisks). Calaxin was accumulated at the ciliary base (white arrowheads). (D) twister-/-. Ciliary localization of Dnah8 was lost (white asterisks). Scale bar: 20 μm.
|
|
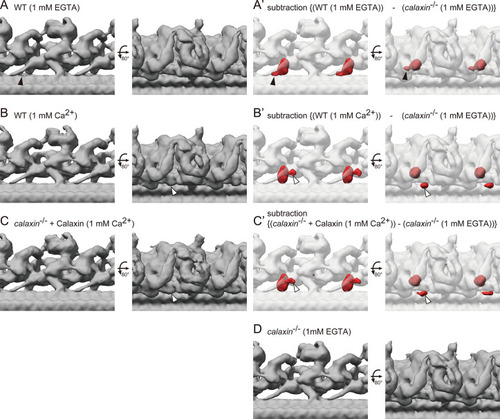
(A–B) OAD-DC structures of WT sperm flagella in different Ca2+ conditions: (A) 1 mM EGTA condition (for Ca2+-free) and (B) 1 mM Ca2+ condition. (C) OAD-DC structure of calaxin-/- sperm flagella incubated with recombinant Calaxin protein in 1 mM Ca2+ condition. (A’-C’) Composite images of difference map (red; subtraction of calaxin-/- in 1 mM EGTA condition from A, B, and C, respectively) and calaxin-/- OAD-DC (translucent). Black arrowheads in A indicate the CCDC151/114 structure adjacent to Calaxin, which is not observed in B and C. White arrowheads in B and C indicate the additional density around DC, which is not observed in A. (D) OAD-DC structure of calaxin-/- sperm in 1 mM EGTA condition, which was used to generate (A’-C’).
|