- Title
-
Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (ehhadh) is essential for production of DHA in zebrafish
- Authors
- Yang, G., Sun, S., He, J., Wang, Y., Ren, T., He, H., Gao, J.
- Source
- Full text @ J. Lipid Res.
|
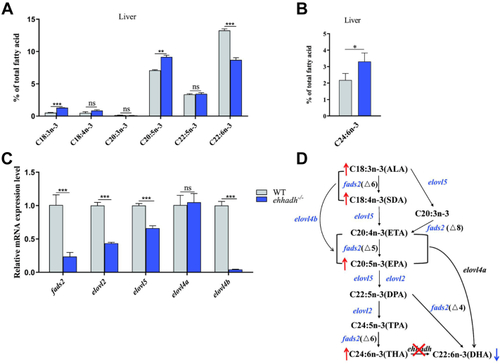
Changes of liver fatty acid compositions and mRNA expression levels of fatty acid metabolism-related genes in Ehhadh-knockout zebrafish (Ehhadh−/−). A: liver n-3PUFA (18:3n-3; 18:4n-3; 20:3n-3; 20:5n-3; 22:5n-3; 22:6n-3) composition of WT zebrafish and Ehhadh−/−. B: liver C24:6n-3 content of WT and Ehhadh−/−. C: the mRNA expression levels of fads2, elovl2, elovl5, elovl4a, and elovl4b in livers of WT and Ehhadh−/−. D: the effect of Ehhadh knockout on DHA synthesis pathway. The blue font indicates that gene expression is significantly downregulated, the blue arrow indicates that the content of related fatty acids is significantly reduced, and the red arrow indicates that the content of related fatty acids is significantly increased. ns means no significant difference; ∗ means P < 0.05; ∗∗ means P < 0.01; ∗∗∗ means P < 0.001. Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; elovl2, fatty acid elongase 2; elovl4a, fatty acid elongase 4a; elovl4b, fatty acid elongase 4b; elovl5, fatty acid elongase 5; fads2, fatty acid desaturase 2; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid. |
|
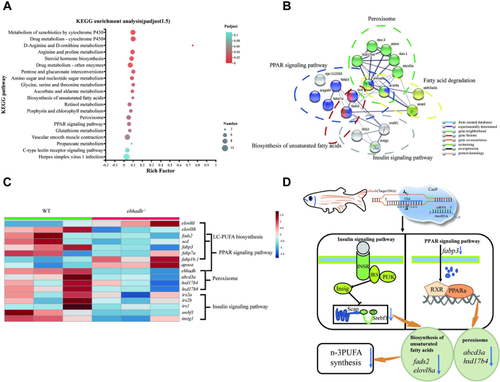
Liver transcriptome analyses of WT zebrafish and Ehhadh-knockout zebrafish (Ehhadh−/−). A: KEGG pathway enrichment analysis of DEG from WT and Ehhadh−/−. B: Network interaction analysis between DEG corresponding proteins. C: DEG heat map analysis related to fatty acid metabolism. D: Flow chart of Ehhadh regulating DHA synthesis through the PPARα and insulin signaling pathways. The blue arrow indicates a significant decrease in expression. DEG, differentially expressed gene; Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase. |
|
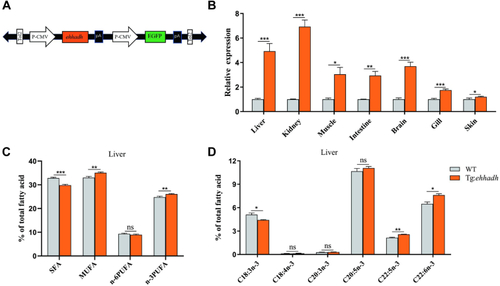
The effect of Ehhadh overexpression on fatty acid composition of zebrafish. A: Schematic depicting of the recombinant plasmid containing the Ehhadh transgene. The Ehhadh gene was driven by the CMV promoter and conjugated to the DNA sequences of Tol2 and GFP fluorescent protein. B: Quantitative expression of Ehhadh in different tissues of WT and Ehhadh transgenic zebrafish (Tg:Ehhadh). C: Liver ∑SFA, ∑MUFA, ∑n-6PUFA, and ∑n-3PUFA composition of WT and Tg:Ehhadh. D: Liver n-3PUFA (18:3n-3, 18:4n-3, 20:3n-3, 20:5n-3, 22:5n-3, and 22:6n-3) composition of WT and Tg:Ehhadh. ns means no significant difference; ∗ means P < 0.05; ∗∗ means P < 0.01; ∗∗∗ means P < 0.001. Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid. |
|
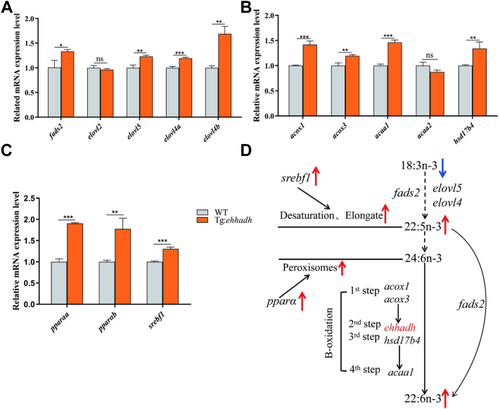
The mRNA expression levels of fatty acid metabolism-related genes in liver of WT and Ehhadh transgenic zebrafish (Tg:Ehhadh). A: Effects of Ehhadh overexpression on mRNA expression of PUFA synthesis-related genes in liver. B: Effects of Ehhadh overexpression on mRNA expression of Peroxisome β-oxidation related genes in liver. C: Effects of Ehhadh overexpression on mRNA expression of pparα and srebf1 in liver. D: The effect of Ehhadh overexpression on DHA synthesis pathway. The blue arrow indicates the downward adjustment, and the red arrow indicates the upward adjustment. ns means no significant difference; ∗ means P < 0.05; ∗∗ means P < 0.01; ∗∗∗ means P < 0.001. acaa1, acetyl-CoA acyltransferase 1; acaa2, acetyl-CoA acyltransferase 2; acox1, acyl-CoA oxidase 1; acox3, acyl-CoA oxidase 1; Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; elovl2, fatty acid elongase 2; elovl4a, fatty acid elongase 4a; elovl4b, fatty acid elongase 4b; elovl5, fatty acid elongase 5; fads2, fatty acid desaturase 2; hsd17b4, hydroxysteroid 17-beta dehydrogenase 4; pparαa, peroxisome proliferator activated receptor alpha a; pparαb, peroxisome proliferator activated receptor alpha b; srebf1, sterol regulatory element binding transcription factor 1. |
|
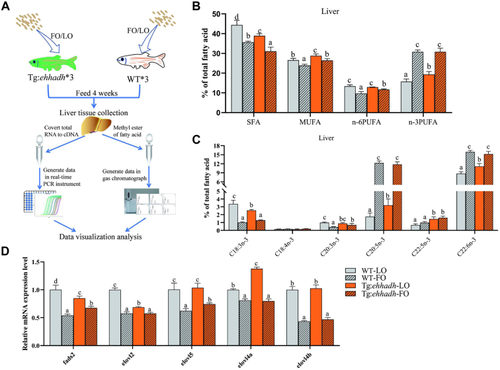
Fatty acid composition of liver and mRNA expression levels of fatty acid metabolism-related genes of zebrafish (WT) and Ehhadh transgenic zebrafish (Tg:Ehhadh) fed with linseed oil (LO) or fish oil (FO) diet for 4 weeks. A: The flow chart of feeding experiment. B: Liver ∑SFA, ∑MUFA, ∑n-6PUFA, and ∑n-3PUFA composition of WT and Tg:Ehhadh. C: Liver fatty acid n-3PUFA compositions of WT and Tg:Ehhadh. D: The mRNA expression levels of fads2, elovl2, elovl5, elovl4a, and elovl4b in livers of WT and Tg:Ehhadh. The different letters above the bars indicated significant difference (P < 0.05). Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; elovl2, fatty acid elongase 2; elovl4a, fatty acid elongase 4a; elovl4b, fatty acid elongase 4b; elovl5, fatty acid elongase 5; fads2, fatty acid desaturase 2; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid. |