- Title
-
In situ assessment of statins' effect on autophagic activity in zebrafish larvae cardiomyocytes
- Authors
- Zhang, J., Zuo, Z., Li, J., Wang, Y., Huang, J., Xu, L., Jin, K., Lu, H., Dai, Y.
- Source
- Full text @ Front Cardiovasc Med
|
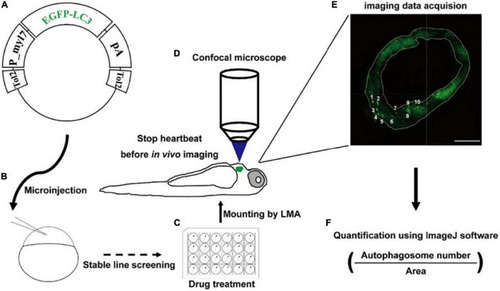
Strategy of monitoring autophagic activity in zebrafish cardiomyocytes |
|
The density of autophagosomes in zebrafish heart is regulated by known autophagy-modulating drugs. |
|
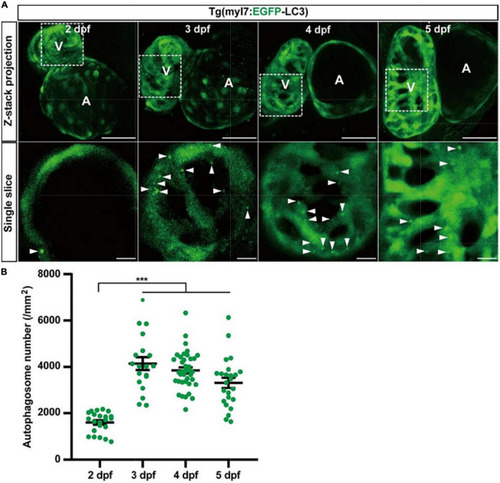
The density of autophagosomes in zebrafish cardiomyocytes is quantified during 2–5 dpf. |
|
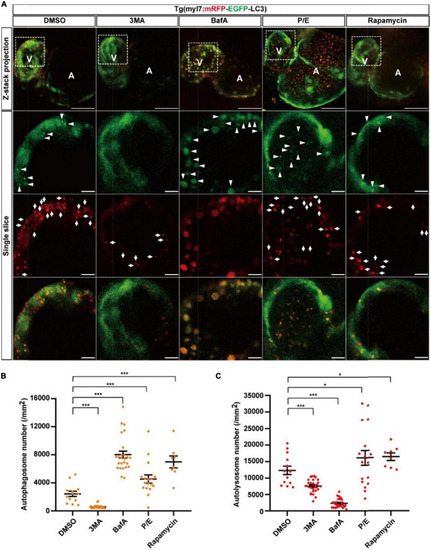
The densities of autophagosomes and autolysosomes in zebrafish cardiomyocytes are regulated by known autophagy-modulating drugs. |
|
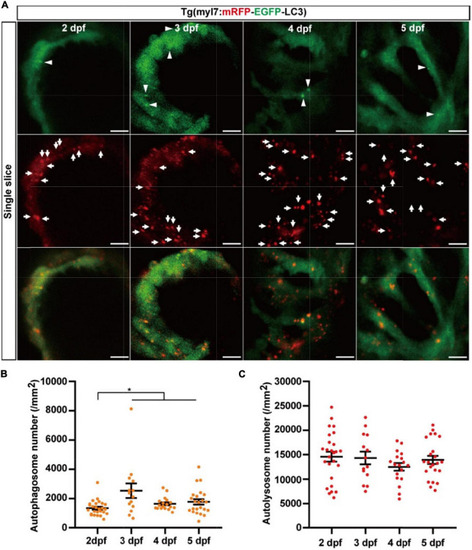
The densities of autophagosomes and autolysosomes in zebrafish hearts are quantified simultaneously during 2–5 dpf. |
|
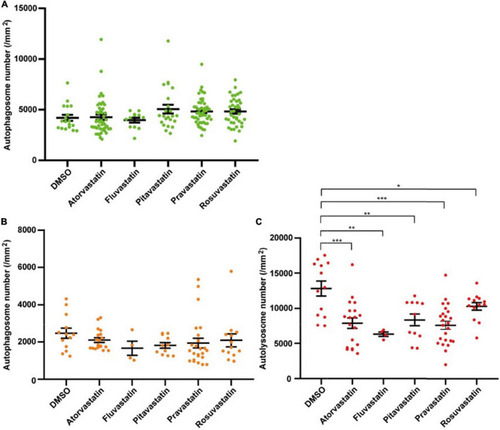
The densities of autophagosomes and autolysosomes in zebrafish cardiomyocytes are quantified when treated with statins. |