- Title
-
Unraveling Differential Transcriptomes and Cell Types in Zebrafish Larvae Intestine and Liver
- Authors
- Gao, Y., Jin, Q., Gao, C., Chen, Y., Sun, Z., Guo, G., Peng, J.
- Source
- Full text @ Cells
|
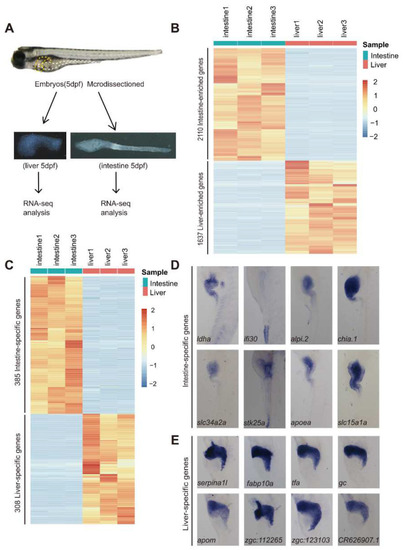
Identification of DEGs between the 5 dpf larval intestine and liver by RNA-seq analysis. ( |
|
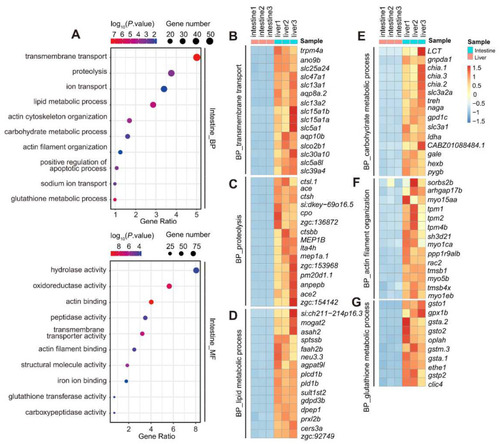
GO analysis of the 2110 intestine-enriched DEGs identified at 5 dpf. ( |
|
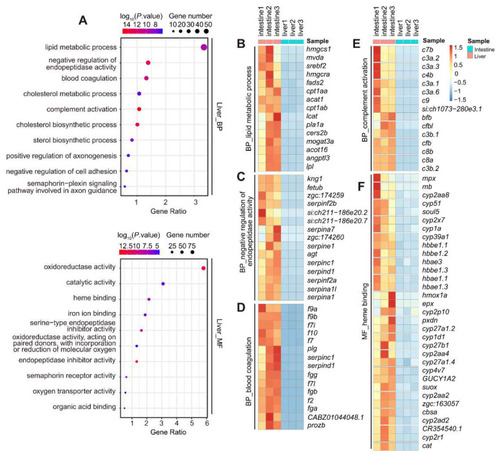
GO analysis of the 1637 liver-enriched DEGs identified at 5dpf. ( |
|
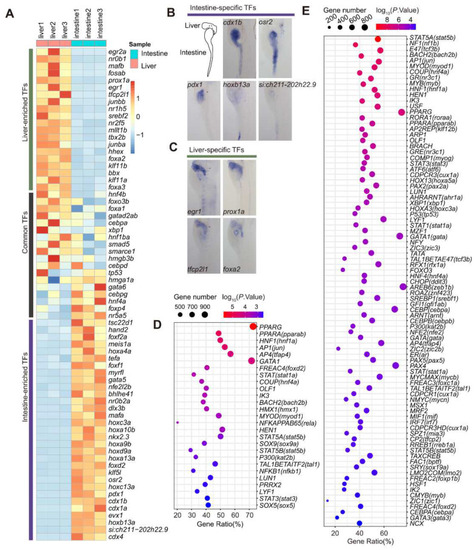
Mining TFs regulating the expression of intestine- and liver-enriched genes at 5 dpf. ( |
|
RNA-seq analysis of the gene expression profiles in the anterior (S1), middle (S2) and posterior regions (S3) of the intestine at 5 dpf. ( |
|
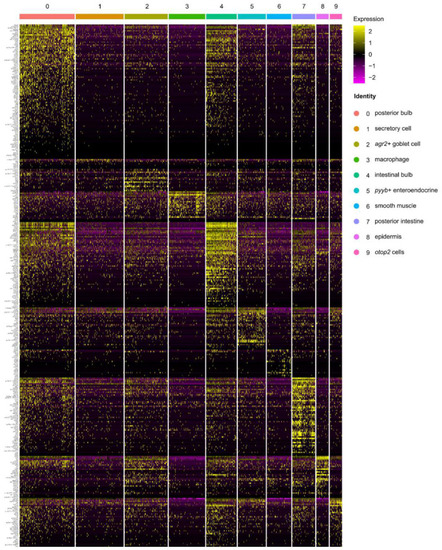
scRNA-seq analysis identified 10 cell types in the WT intestine at 5 dpf. ( |
|
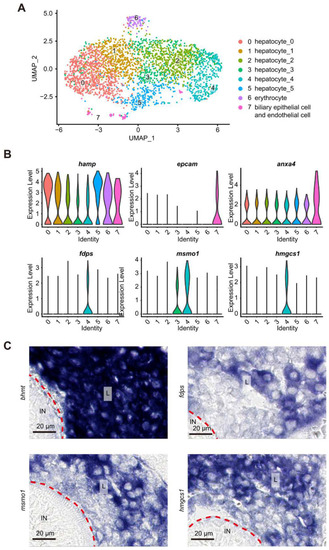
scRNA-seq analysis identified 8 cell clusters in the WT liver at 5 dpf. ( |
|
Cellular expression specificities of the 385 intestine-specific genes. Heatmap showing the expression of the 375 intestine-specific genes (10 genes were not detected by the intestine scRNA-seq) at the single-cell level in each cell cluster identified by the intestine scRNA-seq. |