- Title
-
Evolutionary and Expression Analysis of MOV10 and MOV10L1 Reveals Their Origin, Duplication and Divergence
- Authors
- Yang, S., Zhang, X., Li, X., Yin, X., Teng, L., Ji, G., Li, H.
- Source
- Full text @ Int. J. Mol. Sci.
|
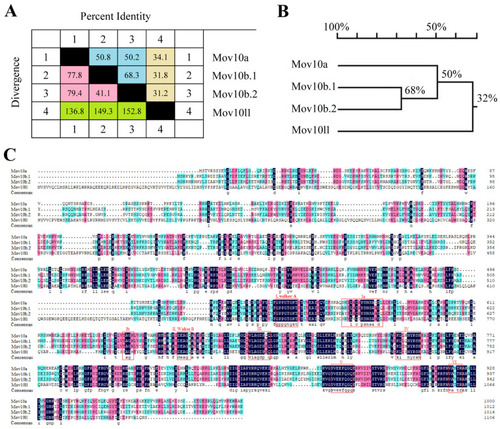
The protein sequences alignment of the Mov10s and Mov10l1 in zebrafish. ( |
|
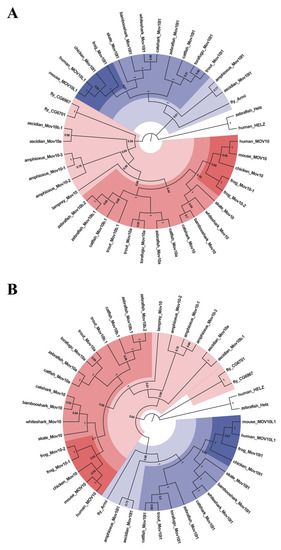
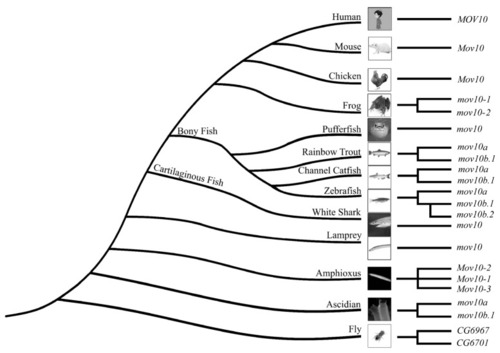
Phylogenetic tree analysis of the MOV10 and MOV10L1 in the selected species above. They are constructed by Maximum-Likelihood method ( |
|
The intron-exon structures of the |
|
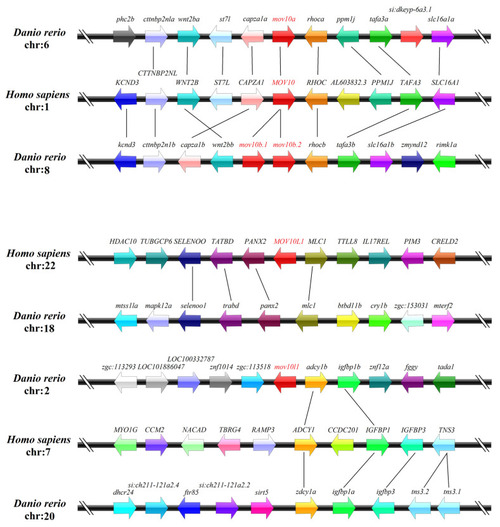
Synteny analyses of the |
|
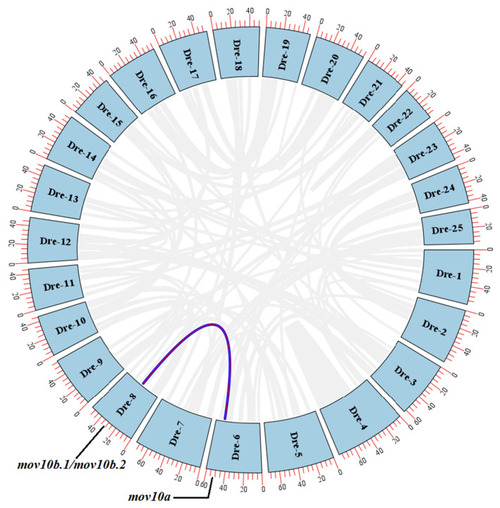
The circos plot of |
|
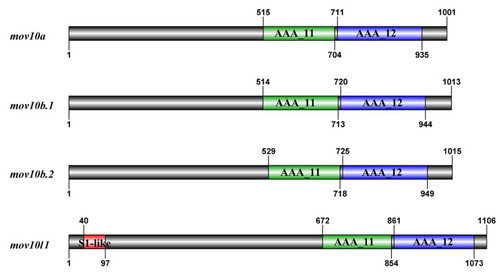
The schematic diagrams of domain organization in zebrafish Mov10s and Mov10l1. The bold black lines mean the amino acid sequence of the proteins. The higher squares represent the predicted domains of Mov10s and Mov10l1. |
|
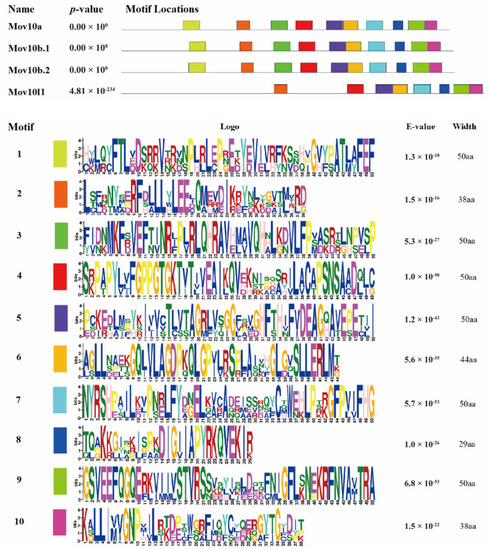
The schematic diagram of motifs of zebrafish Mov10s and Mov10l1 proteins. The motifs are arranged according to the position on the protein sequence, the letters in each motif are amino abbreviation. The size of the letter represents the saliency of the amino acid in the motif. The larger the letter, the higher the saliency, which is, the higher the frequency at which the amino acid appears in the same position in the same motif in different sequences. |
|
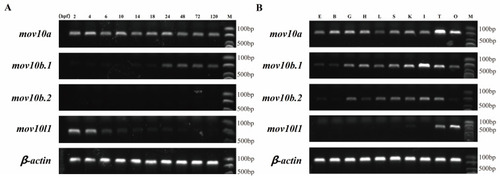
The Expression electrophoretogram of zebrafish |
|
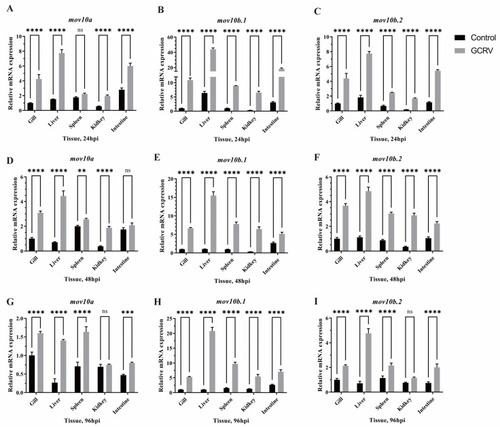
The antiviral properties of |
|
The lineage-specific intra-chromosomally duplications and tandem duplication of mov10 occurred among species. |