- Title
-
Optogenetic control of YAP cellular localisation and function
- Authors
- Toh, P.J.Y., Lai, J.K.H., Hermann, A., Destaing, O., Sheetz, M.P., Sudol, M., Saunders, T.E.
- Source
- Full text @ EMBO Rep.
|
|
|
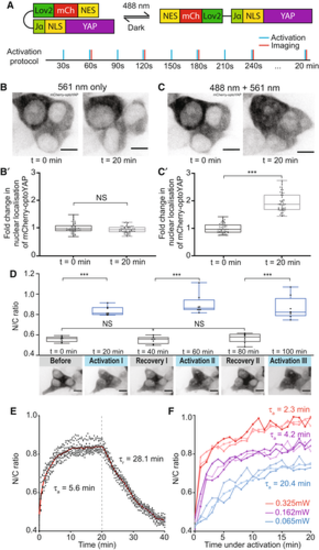
Figure 2. Light activation of optoYAP can activate downstream YAP target genes and control cell proliferation |
|
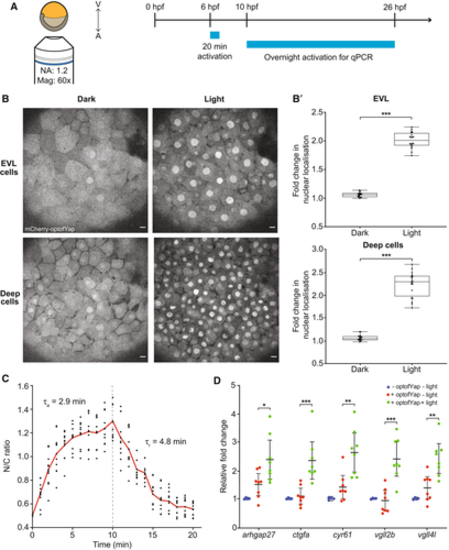
Figure 3. Validation of optofYap in zebrafish |
|
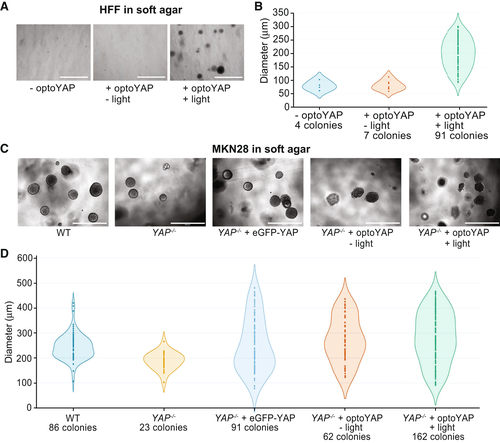
Figure 4. Functional assays of optoYAP in tissue culture cells |